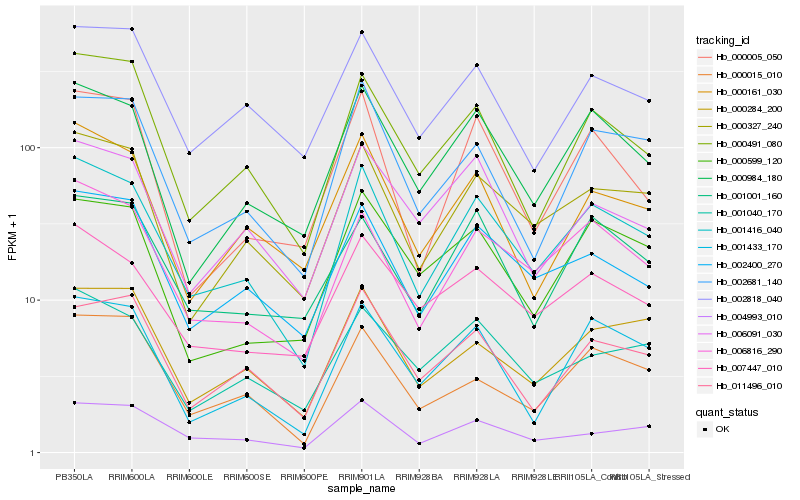
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001416_040 |
0.0 |
- |
- |
acid phosphatase, putative [Ricinus communis] |
| 2 |
Hb_006816_290 |
0.080387025 |
- |
- |
PREDICTED: uncharacterized protein LOC105647449 [Jatropha curcas] |
| 3 |
Hb_000327_240 |
0.0818365623 |
- |
- |
PREDICTED: interaptin-like [Jatropha curcas] |
| 4 |
Hb_000984_180 |
0.0894511863 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002400_270 |
0.0949344 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000491_080 |
0.1020446696 |
- |
- |
PREDICTED: F-box protein SKIP31 [Jatropha curcas] |
| 7 |
Hb_001433_170 |
0.1027361809 |
- |
- |
PREDICTED: topless-related protein 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000161_030 |
0.1038969171 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP8 [Jatropha curcas] |
| 9 |
Hb_007447_010 |
0.1045874649 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB3 isoform X3 [Jatropha curcas] |
| 10 |
Hb_004993_010 |
0.1054457642 |
- |
- |
PREDICTED: uncharacterized protein LOC104889591 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_002681_140 |
0.1077334831 |
- |
- |
PREDICTED: surfeit locus protein 2 [Jatropha curcas] |
| 12 |
Hb_001040_170 |
0.1083803141 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21705, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_002818_040 |
0.1134402538 |
- |
- |
tRNA synthetase-related family protein [Populus trichocarpa] |
| 14 |
Hb_000015_010 |
0.1149194293 |
- |
- |
- |
| 15 |
Hb_000005_050 |
0.1156338297 |
- |
- |
PREDICTED: uncharacterized protein LOC105627993 [Jatropha curcas] |
| 16 |
Hb_000599_120 |
0.1160044092 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT34 [Jatropha curcas] |
| 17 |
Hb_001001_160 |
0.1170453389 |
- |
- |
Phosphatidic acid phosphohydrolase 2 isoform 1 [Theobroma cacao] |
| 18 |
Hb_000284_200 |
0.1174472444 |
- |
- |
- |
| 19 |
Hb_006091_030 |
0.1176218738 |
desease resistance |
Gene Name: AAA |
PREDICTED: cell division control protein 48 homolog C-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_011496_010 |
0.1188243642 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g38420, mitochondrial [Jatropha curcas] |