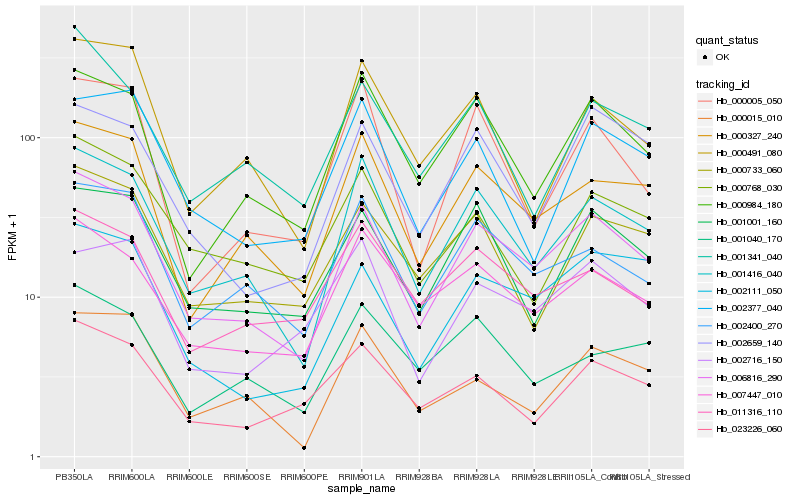
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006816_290 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647449 [Jatropha curcas] |
| 2 |
Hb_001416_040 |
0.080387025 |
- |
- |
acid phosphatase, putative [Ricinus communis] |
| 3 |
Hb_000327_240 |
0.0986053974 |
- |
- |
PREDICTED: interaptin-like [Jatropha curcas] |
| 4 |
Hb_002400_270 |
0.1048016047 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002111_050 |
0.1118850694 |
- |
- |
PREDICTED: uncharacterized protein LOC105641242 [Jatropha curcas] |
| 6 |
Hb_001001_160 |
0.1131228925 |
- |
- |
Phosphatidic acid phosphohydrolase 2 isoform 1 [Theobroma cacao] |
| 7 |
Hb_007447_010 |
0.113282054 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB3 isoform X3 [Jatropha curcas] |
| 8 |
Hb_000984_180 |
0.1141158444 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000733_060 |
0.1206743754 |
- |
- |
PREDICTED: protein GUCD1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002659_140 |
0.1254159718 |
- |
- |
hypothetical protein JCGZ_08031 [Jatropha curcas] |
| 11 |
Hb_000768_030 |
0.1254746423 |
- |
- |
PREDICTED: ELMO domain-containing protein A isoform X1 [Jatropha curcas] |
| 12 |
Hb_023226_060 |
0.1266518781 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_011316_110 |
0.1274436968 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At4g25390 [Jatropha curcas] |
| 14 |
Hb_000015_010 |
0.131370339 |
- |
- |
- |
| 15 |
Hb_000491_080 |
0.1316474907 |
- |
- |
PREDICTED: F-box protein SKIP31 [Jatropha curcas] |
| 16 |
Hb_001341_040 |
0.1329196506 |
- |
- |
hypothetical protein JCGZ_05578 [Jatropha curcas] |
| 17 |
Hb_002377_040 |
0.134408234 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 18 |
Hb_002716_150 |
0.1351929125 |
- |
- |
starch synthase isoform I [Manihot esculenta] |
| 19 |
Hb_000005_050 |
0.1362609065 |
- |
- |
PREDICTED: uncharacterized protein LOC105627993 [Jatropha curcas] |
| 20 |
Hb_001040_170 |
0.1375404914 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21705, mitochondrial isoform X1 [Jatropha curcas] |