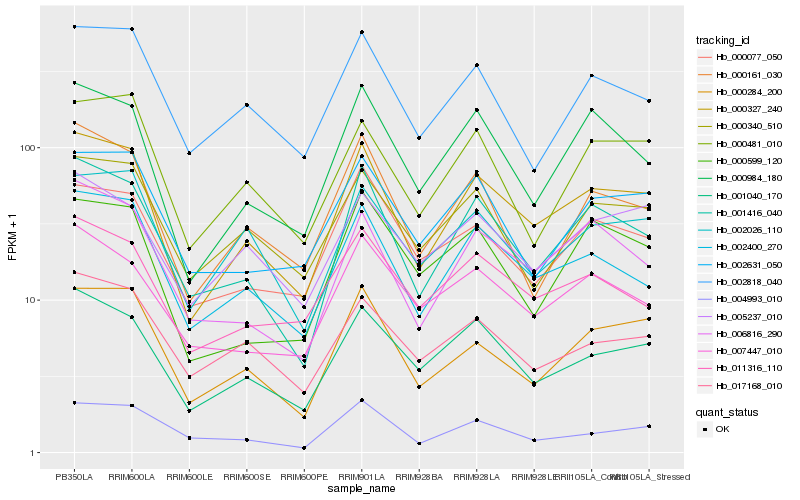
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000327_240 |
0.0 |
- |
- |
PREDICTED: interaptin-like [Jatropha curcas] |
| 2 |
Hb_001416_040 |
0.0818365623 |
- |
- |
acid phosphatase, putative [Ricinus communis] |
| 3 |
Hb_002400_270 |
0.0828496026 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001040_170 |
0.0881881559 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21705, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_000284_200 |
0.0940243136 |
- |
- |
- |
| 6 |
Hb_000984_180 |
0.0978376483 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006816_290 |
0.0986053974 |
- |
- |
PREDICTED: uncharacterized protein LOC105647449 [Jatropha curcas] |
| 8 |
Hb_000161_030 |
0.1046840363 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP8 [Jatropha curcas] |
| 9 |
Hb_002026_110 |
0.1092623995 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_002818_040 |
0.1114489857 |
- |
- |
tRNA synthetase-related family protein [Populus trichocarpa] |
| 11 |
Hb_011316_110 |
0.1114789579 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At4g25390 [Jatropha curcas] |
| 12 |
Hb_002631_050 |
0.1115501529 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 13 |
Hb_000481_010 |
0.1120826585 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP6 [Jatropha curcas] |
| 14 |
Hb_004993_010 |
0.113595864 |
- |
- |
PREDICTED: uncharacterized protein LOC104889591 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_000340_510 |
0.1142071576 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL1 [Jatropha curcas] |
| 16 |
Hb_000077_050 |
0.1164847573 |
- |
- |
PREDICTED: elongation factor G-1, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_017168_010 |
0.1166611292 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007447_010 |
0.1178619027 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB3 isoform X3 [Jatropha curcas] |
| 19 |
Hb_000599_120 |
0.119276618 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT34 [Jatropha curcas] |
| 20 |
Hb_005237_010 |
0.1196337999 |
- |
- |
PREDICTED: uncharacterized protein LOC105630921 [Jatropha curcas] |