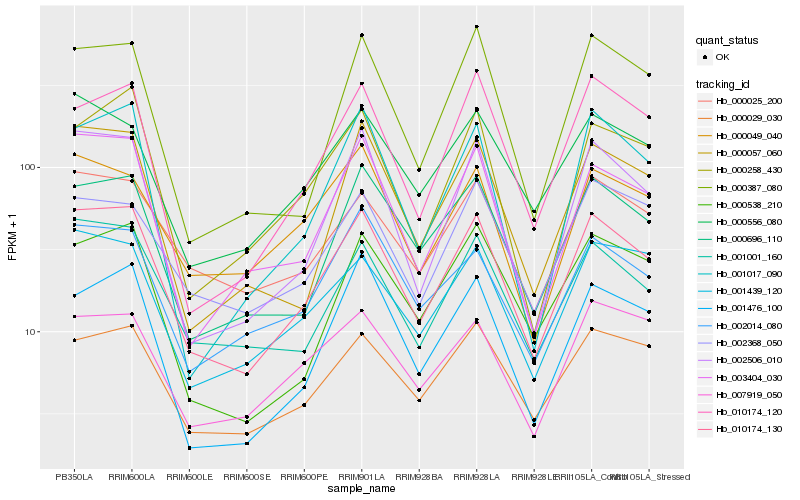
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010174_130 |
0.0 |
- |
- |
PREDICTED: regulator of G-protein signaling 1 [Jatropha curcas] |
| 2 |
Hb_001001_160 |
0.0805359028 |
- |
- |
Phosphatidic acid phosphohydrolase 2 isoform 1 [Theobroma cacao] |
| 3 |
Hb_002506_010 |
0.081049086 |
- |
- |
choline transporter-related family protein [Populus trichocarpa] |
| 4 |
Hb_000696_110 |
0.0834995537 |
- |
- |
PREDICTED: la-related protein 1C-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_003404_030 |
0.0838748677 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase 2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000029_030 |
0.0892123767 |
- |
- |
PREDICTED: mitochondrial fission protein ELM1 [Jatropha curcas] |
| 7 |
Hb_000025_200 |
0.0921336939 |
- |
- |
PREDICTED: cell division control protein 2 homolog A isoform X1 [Jatropha curcas] |
| 8 |
Hb_000057_060 |
0.0932480027 |
- |
- |
PREDICTED: uncharacterized protein LOC105634990 isoform X4 [Jatropha curcas] |
| 9 |
Hb_001439_120 |
0.0939627397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000556_080 |
0.0939638537 |
- |
- |
PREDICTED: synaptotagmin-5 [Pyrus x bretschneideri] |
| 11 |
Hb_002014_080 |
0.0974092827 |
- |
- |
PREDICTED: tankyrase-1 isoform X4 [Jatropha curcas] |
| 12 |
Hb_000258_430 |
0.0989828382 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 13 |
Hb_010174_120 |
0.0996660098 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1-like [Gossypium raimondii] |
| 14 |
Hb_000049_040 |
0.1005667845 |
- |
- |
PREDICTED: uncharacterized protein LOC105644475 [Jatropha curcas] |
| 15 |
Hb_000538_210 |
0.1030441095 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_000387_080 |
0.1030951909 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002368_050 |
0.1033946765 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007919_050 |
0.1037051125 |
- |
- |
PREDICTED: uncharacterized protein LOC105635142 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001476_100 |
0.1037405274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001017_090 |
0.1039992928 |
- |
- |
- |