| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
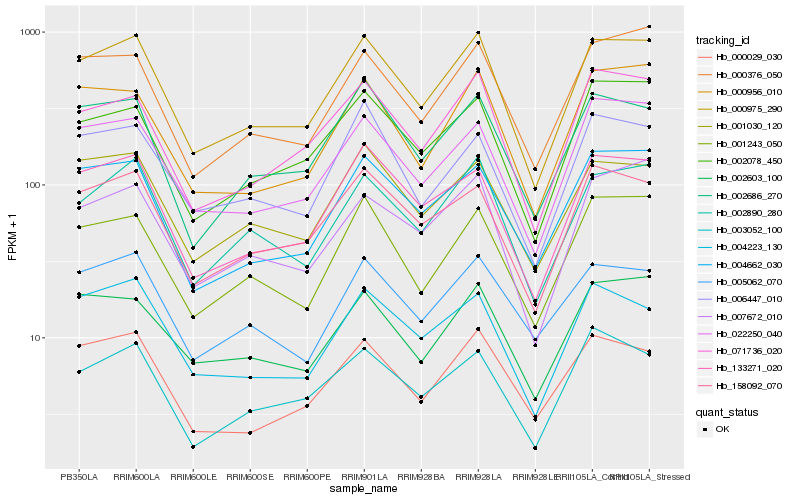
Hb_000975_290 |
0.0 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 2 |
Hb_002890_280 |
0.0606453365 |
- |
- |
hypothetical protein PRUPE_ppa009665mg [Prunus persica] |
| 3 |
Hb_133271_020 |
0.0617065484 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 4 |
Hb_158092_070 |
0.0649905805 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_001030_120 |
0.0677743086 |
- |
- |
PREDICTED: tyrosine--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 6 |
Hb_002686_270 |
0.0686367487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_071736_020 |
0.0699323359 |
- |
- |
hypothetical protein POPTR_0006s10330g [Populus trichocarpa] |
| 8 |
Hb_004662_030 |
0.0706975584 |
- |
- |
hypothetical protein PRUPE_ppa012487mg [Prunus persica] |
| 9 |
Hb_022250_040 |
0.0712150712 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 2, mitochondrial [Jatropha curcas] |
| 10 |
Hb_001243_050 |
0.0714444041 |
- |
- |
PREDICTED: uncharacterized protein LOC105643776 [Jatropha curcas] |
| 11 |
Hb_004223_130 |
0.0744196722 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002078_450 |
0.0749068526 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 13 |
Hb_000376_050 |
0.0758963822 |
- |
- |
polyadenylate-binding protein, putative [Ricinus communis] |
| 14 |
Hb_000029_030 |
0.0759548386 |
- |
- |
PREDICTED: mitochondrial fission protein ELM1 [Jatropha curcas] |
| 15 |
Hb_002603_100 |
0.076329325 |
- |
- |
ATATH9, putative [Ricinus communis] |
| 16 |
Hb_000956_010 |
0.0787427743 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like [Jatropha curcas] |
| 17 |
Hb_006447_010 |
0.0813966637 |
- |
- |
PREDICTED: peroxisome biogenesis protein 22 [Jatropha curcas] |
| 18 |
Hb_005062_070 |
0.0815054034 |
- |
- |
RNA splicing protein mrs2, mitochondrial, putative [Ricinus communis] |
| 19 |
Hb_003052_100 |
0.0816870267 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_007672_010 |
0.0817462741 |
- |
- |
PREDICTED: uncharacterized protein LOC105639669 isoform X1 [Jatropha curcas] |