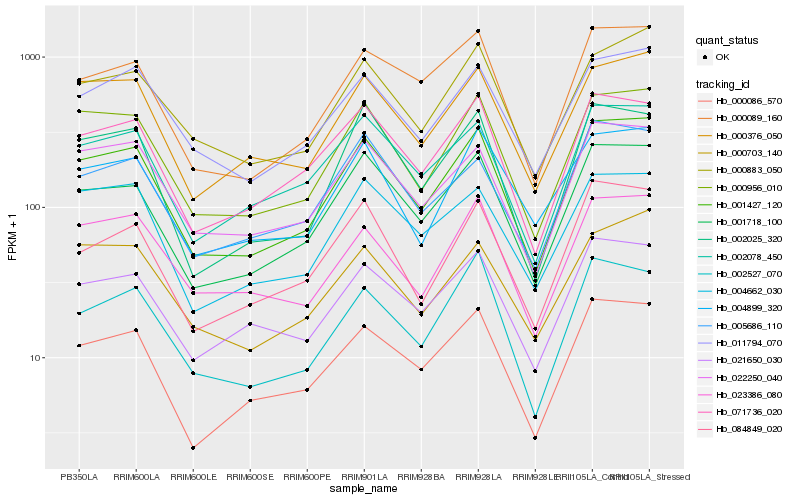
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001427_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637787 [Jatropha curcas] |
| 2 |
Hb_000956_010 |
0.0465913496 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like [Jatropha curcas] |
| 3 |
Hb_001718_100 |
0.0493895745 |
- |
- |
PREDICTED: casein kinase II subunit beta-like isoform X5 [Jatropha curcas] |
| 4 |
Hb_000086_570 |
0.0615228424 |
- |
- |
hypothetical protein JCGZ_24934 [Jatropha curcas] |
| 5 |
Hb_000376_050 |
0.0615465195 |
- |
- |
polyadenylate-binding protein, putative [Ricinus communis] |
| 6 |
Hb_071736_020 |
0.0644257337 |
- |
- |
hypothetical protein POPTR_0006s10330g [Populus trichocarpa] |
| 7 |
Hb_022250_040 |
0.0645828846 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 2, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000089_160 |
0.0674173387 |
- |
- |
60S ribosomal protein L5B [Hevea brasiliensis] |
| 9 |
Hb_011794_070 |
0.0683005016 |
- |
- |
PREDICTED: 60S ribosomal protein L36-2-like [Jatropha curcas] |
| 10 |
Hb_002078_450 |
0.0683512348 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 11 |
Hb_000883_050 |
0.0683726668 |
- |
- |
PREDICTED: 60S ribosomal protein L37a-like, partial [Solanum tuberosum] |
| 12 |
Hb_002025_320 |
0.0725197201 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit I-like [Jatropha curcas] |
| 13 |
Hb_084849_020 |
0.0742680979 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 14 |
Hb_021650_030 |
0.0744090263 |
- |
- |
PREDICTED: uncharacterized protein LOC105642056 [Jatropha curcas] |
| 15 |
Hb_023386_080 |
0.0757646015 |
- |
- |
PREDICTED: peroxisome biogenesis protein 19-2-like [Jatropha curcas] |
| 16 |
Hb_002527_070 |
0.0759674953 |
- |
- |
axonemal dynein light chain, putative [Ricinus communis] |
| 17 |
Hb_004662_030 |
0.07648549 |
- |
- |
hypothetical protein PRUPE_ppa012487mg [Prunus persica] |
| 18 |
Hb_004899_320 |
0.0771407811 |
- |
- |
PREDICTED: oligouridylate-binding protein 1-like [Jatropha curcas] |
| 19 |
Hb_005686_110 |
0.0788707861 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Populus euphratica] |
| 20 |
Hb_000703_140 |
0.0790058995 |
- |
- |
transcription initiation factor TFIID subunit 13-like [Jatropha curcas] |