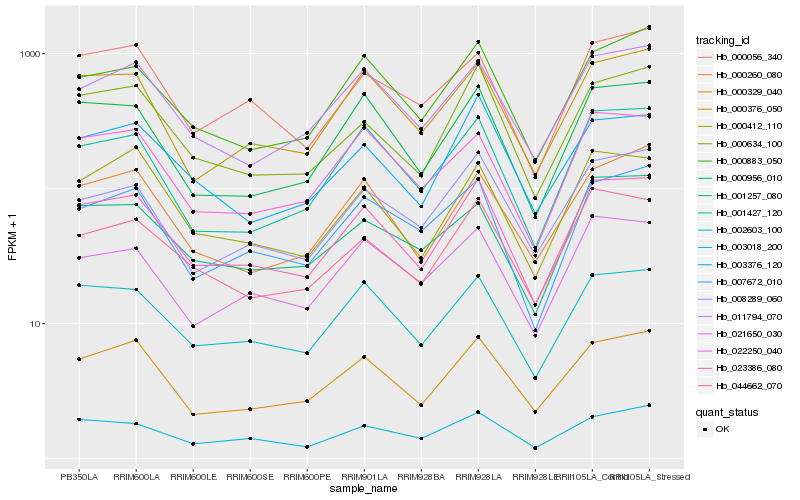
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023386_080 |
0.0 |
- |
- |
PREDICTED: peroxisome biogenesis protein 19-2-like [Jatropha curcas] |
| 2 |
Hb_002603_100 |
0.0550689564 |
- |
- |
ATATH9, putative [Ricinus communis] |
| 3 |
Hb_000329_040 |
0.0590677165 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3 [Jatropha curcas] |
| 4 |
Hb_000634_100 |
0.0665596506 |
- |
- |
hypothetical protein PRUPE_ppa013350mg [Prunus persica] |
| 5 |
Hb_044662_070 |
0.0676449598 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3 [Jatropha curcas] |
| 6 |
Hb_000956_010 |
0.0678683753 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like [Jatropha curcas] |
| 7 |
Hb_000376_050 |
0.0702667178 |
- |
- |
polyadenylate-binding protein, putative [Ricinus communis] |
| 8 |
Hb_011794_070 |
0.0726809216 |
- |
- |
PREDICTED: 60S ribosomal protein L36-2-like [Jatropha curcas] |
| 9 |
Hb_003018_200 |
0.0753422378 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 10 |
Hb_001427_120 |
0.0757646015 |
- |
- |
PREDICTED: uncharacterized protein LOC105637787 [Jatropha curcas] |
| 11 |
Hb_000883_050 |
0.075848954 |
- |
- |
PREDICTED: 60S ribosomal protein L37a-like, partial [Solanum tuberosum] |
| 12 |
Hb_022250_040 |
0.0759188871 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 2, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000412_110 |
0.077113675 |
- |
- |
Rop4 [Hevea brasiliensis] |
| 14 |
Hb_021650_030 |
0.0782901881 |
- |
- |
PREDICTED: uncharacterized protein LOC105642056 [Jatropha curcas] |
| 15 |
Hb_003376_120 |
0.0788898254 |
- |
- |
PREDICTED: cell number regulator 8 [Jatropha curcas] |
| 16 |
Hb_007672_010 |
0.079319873 |
- |
- |
PREDICTED: uncharacterized protein LOC105639669 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000260_080 |
0.080642254 |
- |
- |
thioredoxin H-type 1 [Hevea brasiliensis] |
| 18 |
Hb_008289_060 |
0.0809994853 |
- |
- |
PREDICTED: general transcription factor IIE subunit 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001257_080 |
0.0820773236 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000056_340 |
0.0832805295 |
- |
- |
60S ribosomal protein L34A [Hevea brasiliensis] |