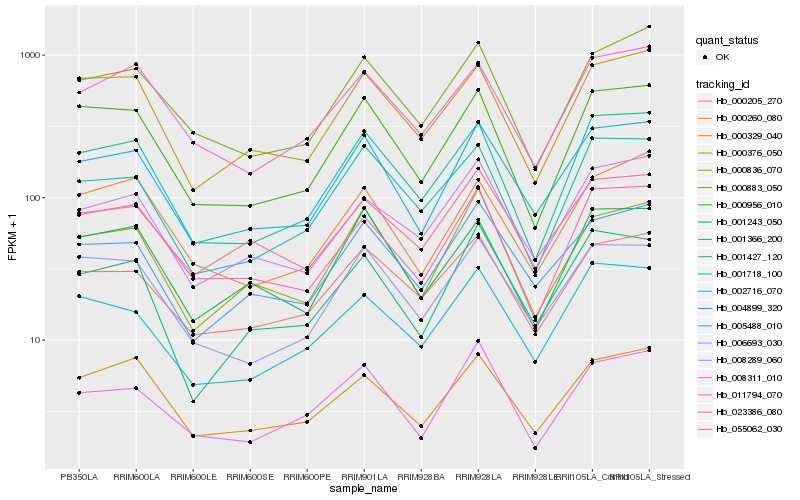
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004899_320 |
0.0 |
- |
- |
PREDICTED: oligouridylate-binding protein 1-like [Jatropha curcas] |
| 2 |
Hb_005488_010 |
0.0569299856 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000836_070 |
0.0673641905 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_006693_030 |
0.0678429487 |
- |
- |
PREDICTED: ribonuclease H2 subunit A [Jatropha curcas] |
| 5 |
Hb_001366_200 |
0.0733695729 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000956_010 |
0.0743136122 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like [Jatropha curcas] |
| 7 |
Hb_000329_040 |
0.074493875 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3 [Jatropha curcas] |
| 8 |
Hb_001427_120 |
0.0771407811 |
- |
- |
PREDICTED: uncharacterized protein LOC105637787 [Jatropha curcas] |
| 9 |
Hb_000205_270 |
0.0780186738 |
- |
- |
PREDICTED: uncharacterized protein LOC105632722 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000376_050 |
0.0781887142 |
- |
- |
polyadenylate-binding protein, putative [Ricinus communis] |
| 11 |
Hb_000883_050 |
0.0795188726 |
- |
- |
PREDICTED: 60S ribosomal protein L37a-like, partial [Solanum tuberosum] |
| 12 |
Hb_008289_060 |
0.0803077175 |
- |
- |
PREDICTED: general transcription factor IIE subunit 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_002716_070 |
0.0810897298 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 14 |
Hb_001243_050 |
0.0820793939 |
- |
- |
PREDICTED: uncharacterized protein LOC105643776 [Jatropha curcas] |
| 15 |
Hb_055062_030 |
0.082282837 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 10b-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_023386_080 |
0.0842721268 |
- |
- |
PREDICTED: peroxisome biogenesis protein 19-2-like [Jatropha curcas] |
| 17 |
Hb_011794_070 |
0.0845515383 |
- |
- |
PREDICTED: 60S ribosomal protein L36-2-like [Jatropha curcas] |
| 18 |
Hb_001718_100 |
0.0857373591 |
- |
- |
PREDICTED: casein kinase II subunit beta-like isoform X5 [Jatropha curcas] |
| 19 |
Hb_008311_010 |
0.0858671603 |
- |
- |
hypothetical protein CARUB_v10003781mg [Capsella rubella] |
| 20 |
Hb_000260_080 |
0.0860939662 |
- |
- |
thioredoxin H-type 1 [Hevea brasiliensis] |