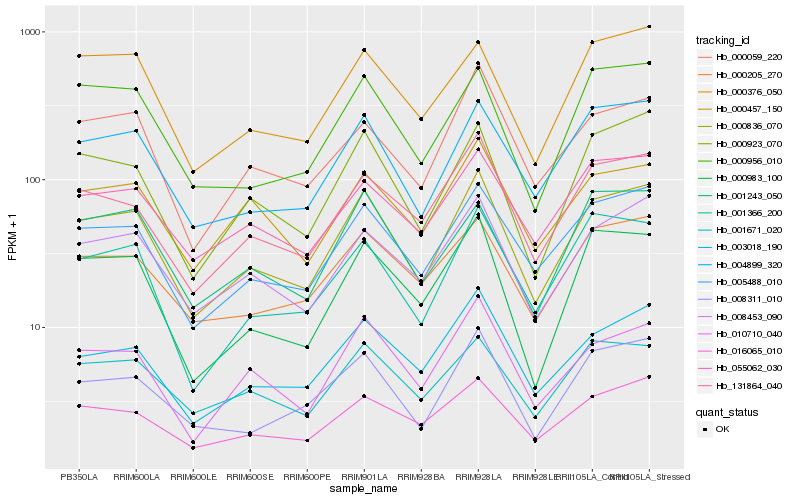
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_070 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_005488_010 |
0.0651008679 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004899_320 |
0.0673641905 |
- |
- |
PREDICTED: oligouridylate-binding protein 1-like [Jatropha curcas] |
| 4 |
Hb_000923_070 |
0.0751684938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_008453_090 |
0.0756425735 |
- |
- |
hypothetical protein JCGZ_08177 [Jatropha curcas] |
| 6 |
Hb_010710_040 |
0.0785986024 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_003018_190 |
0.0805761687 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g31070, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000983_100 |
0.0815435084 |
- |
- |
PREDICTED: AP-4 complex subunit mu isoform X1 [Vitis vinifera] |
| 9 |
Hb_131864_040 |
0.0823216065 |
- |
- |
PREDICTED: uncharacterized protein LOC105649406 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001671_020 |
0.0835736312 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g08510 [Jatropha curcas] |
| 11 |
Hb_008311_010 |
0.0844117739 |
- |
- |
hypothetical protein CARUB_v10003781mg [Capsella rubella] |
| 12 |
Hb_000376_050 |
0.0846987044 |
- |
- |
polyadenylate-binding protein, putative [Ricinus communis] |
| 13 |
Hb_000205_270 |
0.0859201335 |
- |
- |
PREDICTED: uncharacterized protein LOC105632722 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000956_010 |
0.0859851041 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like [Jatropha curcas] |
| 15 |
Hb_001366_200 |
0.0874368845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001243_050 |
0.0876461995 |
- |
- |
PREDICTED: uncharacterized protein LOC105643776 [Jatropha curcas] |
| 17 |
Hb_016065_010 |
0.0879512318 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g08305 [Jatropha curcas] |
| 18 |
Hb_055062_030 |
0.0888362727 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 10b-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000059_220 |
0.0891439981 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |
| 20 |
Hb_000457_150 |
0.0904419102 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |