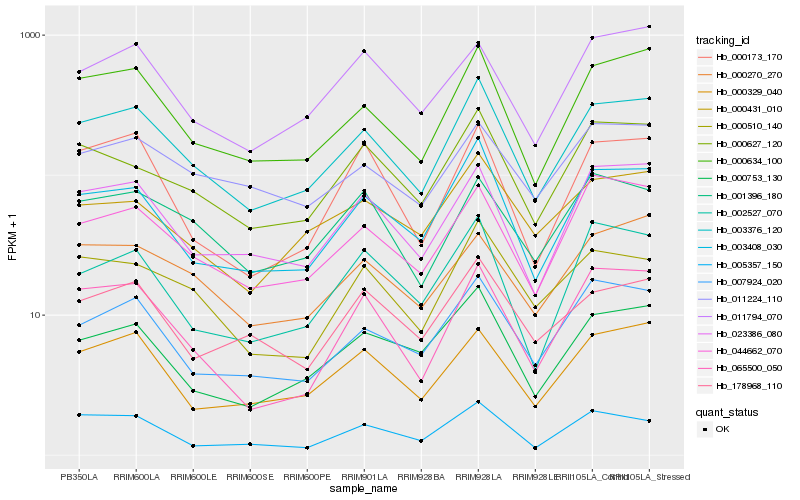
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003376_120 |
0.0 |
- |
- |
PREDICTED: cell number regulator 8 [Jatropha curcas] |
| 2 |
Hb_000634_100 |
0.0624351985 |
- |
- |
hypothetical protein PRUPE_ppa013350mg [Prunus persica] |
| 3 |
Hb_003408_030 |
0.0701321948 |
- |
- |
PREDICTED: protein YIF1B [Jatropha curcas] |
| 4 |
Hb_023386_080 |
0.0788898254 |
- |
- |
PREDICTED: peroxisome biogenesis protein 19-2-like [Jatropha curcas] |
| 5 |
Hb_044662_070 |
0.0859891739 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3 [Jatropha curcas] |
| 6 |
Hb_007924_020 |
0.0875511517 |
- |
- |
PREDICTED: chaperonin CPN60-like 2, mitochondrial isoform X2 [Jatropha curcas] |
| 7 |
Hb_000627_120 |
0.0885563788 |
- |
- |
PREDICTED: probable adenylate kinase 7, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000329_040 |
0.0894301514 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3 [Jatropha curcas] |
| 9 |
Hb_000753_130 |
0.0924297527 |
- |
- |
PREDICTED: uncharacterized protein LOC105640673 [Jatropha curcas] |
| 10 |
Hb_000510_140 |
0.0931330488 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2 [Jatropha curcas] |
| 11 |
Hb_000270_270 |
0.0990558418 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000431_010 |
0.100406732 |
- |
- |
hypothetical protein POPTR_0016s05340g [Populus trichocarpa] |
| 13 |
Hb_065500_050 |
0.1006205437 |
- |
- |
lipoate-protein ligase B, putative [Ricinus communis] |
| 14 |
Hb_001396_180 |
0.1021286117 |
- |
- |
PREDICTED: uncharacterized protein LOC105644834 [Jatropha curcas] |
| 15 |
Hb_011794_070 |
0.1029584049 |
- |
- |
PREDICTED: 60S ribosomal protein L36-2-like [Jatropha curcas] |
| 16 |
Hb_178968_110 |
0.1031871415 |
- |
- |
PREDICTED: uncharacterized protein LOC105630268 [Jatropha curcas] |
| 17 |
Hb_002527_070 |
0.1033036843 |
- |
- |
axonemal dynein light chain, putative [Ricinus communis] |
| 18 |
Hb_005357_150 |
0.1047724307 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000173_170 |
0.105250536 |
- |
- |
PREDICTED: UPF0613 protein PB24D3.06c [Jatropha curcas] |
| 20 |
Hb_011224_110 |
0.1053830201 |
- |
- |
Pre-mRNA-splicing factor cwc15, putative [Ricinus communis] |