| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
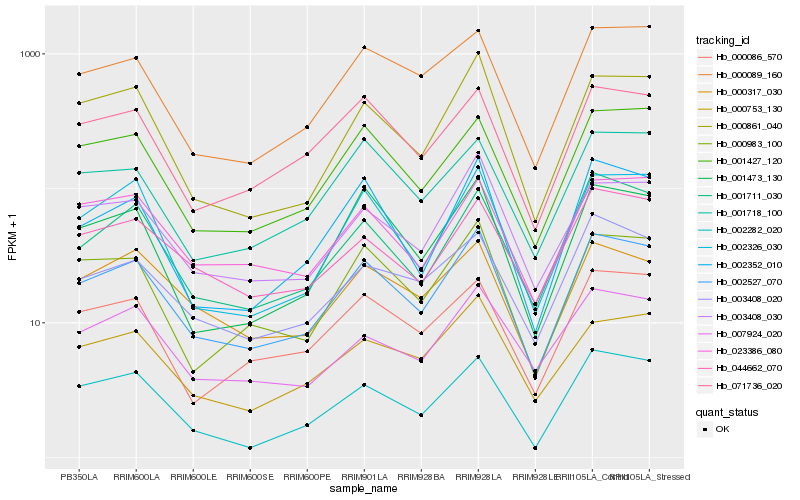
Hb_002527_070 |
0.0 |
- |
- |
axonemal dynein light chain, putative [Ricinus communis] |
| 2 |
Hb_001427_120 |
0.0759674953 |
- |
- |
PREDICTED: uncharacterized protein LOC105637787 [Jatropha curcas] |
| 3 |
Hb_000861_040 |
0.0774419088 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 1 [Jatropha curcas] |
| 4 |
Hb_001711_030 |
0.0776926327 |
- |
- |
PREDICTED: uncharacterized protein LOC105642409 [Jatropha curcas] |
| 5 |
Hb_003408_030 |
0.0777480065 |
- |
- |
PREDICTED: protein YIF1B [Jatropha curcas] |
| 6 |
Hb_000983_100 |
0.0782326989 |
- |
- |
PREDICTED: AP-4 complex subunit mu isoform X1 [Vitis vinifera] |
| 7 |
Hb_002352_010 |
0.0802677742 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 8 |
Hb_071736_020 |
0.0819646171 |
- |
- |
hypothetical protein POPTR_0006s10330g [Populus trichocarpa] |
| 9 |
Hb_001473_130 |
0.0826230048 |
- |
- |
PREDICTED: ubiquinone biosynthesis monooxygenase COQ6 [Jatropha curcas] |
| 10 |
Hb_002282_020 |
0.0830462999 |
- |
- |
PREDICTED: uncharacterized protein LOC105629828 [Jatropha curcas] |
| 11 |
Hb_002326_030 |
0.0832468549 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein REM16-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000089_160 |
0.0838889319 |
- |
- |
60S ribosomal protein L5B [Hevea brasiliensis] |
| 13 |
Hb_000753_130 |
0.0857275721 |
- |
- |
PREDICTED: uncharacterized protein LOC105640673 [Jatropha curcas] |
| 14 |
Hb_023386_080 |
0.0864259197 |
- |
- |
PREDICTED: peroxisome biogenesis protein 19-2-like [Jatropha curcas] |
| 15 |
Hb_000086_570 |
0.0880247022 |
- |
- |
hypothetical protein JCGZ_24934 [Jatropha curcas] |
| 16 |
Hb_003408_020 |
0.0880812361 |
- |
- |
hypothetical protein RCOM_0453040 [Ricinus communis] |
| 17 |
Hb_001718_100 |
0.0920740338 |
- |
- |
PREDICTED: casein kinase II subunit beta-like isoform X5 [Jatropha curcas] |
| 18 |
Hb_044662_070 |
0.0925351175 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3 [Jatropha curcas] |
| 19 |
Hb_000317_030 |
0.0927630165 |
transcription factor |
TF Family: ULT |
PREDICTED: protein ULTRAPETALA 1-like [Jatropha curcas] |
| 20 |
Hb_007924_020 |
0.0931444828 |
- |
- |
PREDICTED: chaperonin CPN60-like 2, mitochondrial isoform X2 [Jatropha curcas] |