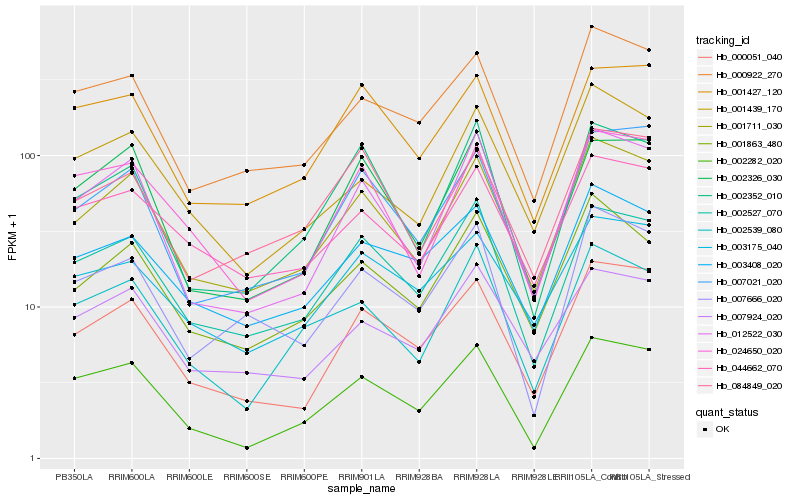
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001711_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642409 [Jatropha curcas] |
| 2 |
Hb_001863_480 |
0.0675018837 |
- |
- |
PREDICTED: uncharacterized protein LOC105630612 [Jatropha curcas] |
| 3 |
Hb_000051_040 |
0.0758318271 |
- |
- |
PREDICTED: uncharacterized protein LOC102597788 [Solanum tuberosum] |
| 4 |
Hb_012522_030 |
0.0761176334 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_002527_070 |
0.0776926327 |
- |
- |
axonemal dynein light chain, putative [Ricinus communis] |
| 6 |
Hb_000922_270 |
0.0805597866 |
- |
- |
iron-sulfur cluster scaffold protein [Hevea brasiliensis] |
| 7 |
Hb_001439_170 |
0.0850174285 |
- |
- |
PREDICTED: pectinesterase 31-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_044662_070 |
0.0864110101 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3 [Jatropha curcas] |
| 9 |
Hb_007021_020 |
0.087848527 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003408_020 |
0.0885989905 |
- |
- |
hypothetical protein RCOM_0453040 [Ricinus communis] |
| 11 |
Hb_007924_020 |
0.0900778302 |
- |
- |
PREDICTED: chaperonin CPN60-like 2, mitochondrial isoform X2 [Jatropha curcas] |
| 12 |
Hb_002539_080 |
0.0921240135 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_084849_020 |
0.0922738091 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 14 |
Hb_002282_020 |
0.0930008843 |
- |
- |
PREDICTED: uncharacterized protein LOC105629828 [Jatropha curcas] |
| 15 |
Hb_002326_030 |
0.0959593958 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein REM16-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_024650_020 |
0.0960708291 |
- |
- |
PREDICTED: uncharacterized protein LOC105646626 [Jatropha curcas] |
| 17 |
Hb_003175_040 |
0.0965384193 |
- |
- |
PREDICTED: katanin p60 ATPase-containing subunit A1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001427_120 |
0.0978905798 |
- |
- |
PREDICTED: uncharacterized protein LOC105637787 [Jatropha curcas] |
| 19 |
Hb_007666_020 |
0.0990374918 |
- |
- |
alkaline phytoceramidase, putative [Ricinus communis] |
| 20 |
Hb_002352_010 |
0.0990625022 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |