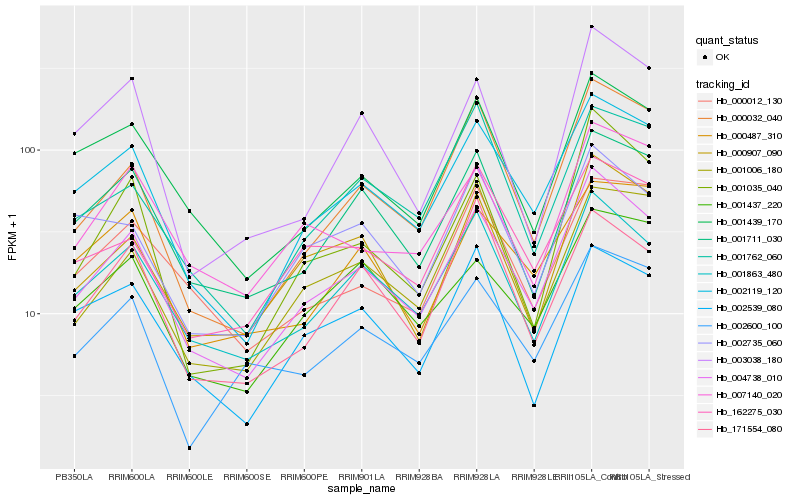
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004738_010 |
0.0 |
- |
- |
PREDICTED: N-acetyl-D-glucosamine kinase-like [Jatropha curcas] |
| 2 |
Hb_002119_120 |
0.0655698606 |
- |
- |
PREDICTED: anthranilate synthase beta subunit 1, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_001863_480 |
0.0797434384 |
- |
- |
PREDICTED: uncharacterized protein LOC105630612 [Jatropha curcas] |
| 4 |
Hb_000907_090 |
0.0964812509 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_001439_170 |
0.099568417 |
- |
- |
PREDICTED: pectinesterase 31-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_001711_030 |
0.1022581081 |
- |
- |
PREDICTED: uncharacterized protein LOC105642409 [Jatropha curcas] |
| 7 |
Hb_000032_040 |
0.1032605774 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 8 |
Hb_001035_040 |
0.1119127214 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_162275_030 |
0.112739505 |
- |
- |
PREDICTED: uncharacterized protein LOC105646638 [Jatropha curcas] |
| 10 |
Hb_007140_020 |
0.1129640067 |
- |
- |
hypothetical protein CICLE_v10008419mg [Citrus clementina] |
| 11 |
Hb_002600_100 |
0.1141672426 |
- |
- |
PREDICTED: putative endo-1,3(4)-beta-glucanase 2 [Jatropha curcas] |
| 12 |
Hb_001437_220 |
0.1177772787 |
- |
- |
Transmembrane protein, putative [Ricinus communis] |
| 13 |
Hb_003038_180 |
0.1198822031 |
- |
- |
PREDICTED: uncharacterized protein LOC105649131 [Jatropha curcas] |
| 14 |
Hb_002735_060 |
0.1200979618 |
- |
- |
PREDICTED: desumoylating isopeptidase 1 [Jatropha curcas] |
| 15 |
Hb_001762_060 |
0.1213383265 |
- |
- |
pantothenate kinase, putative [Ricinus communis] |
| 16 |
Hb_000012_130 |
0.1243927283 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |
| 17 |
Hb_001006_180 |
0.1257776013 |
- |
- |
PREDICTED: endonuclease V-like [Jatropha curcas] |
| 18 |
Hb_000487_310 |
0.1273914673 |
- |
- |
PREDICTED: uncharacterized protein LOC105646211 isoform X1 [Jatropha curcas] |
| 19 |
Hb_171554_080 |
0.1278337057 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 20 |
Hb_002539_080 |
0.128471568 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator, chloroplastic-like [Jatropha curcas] |