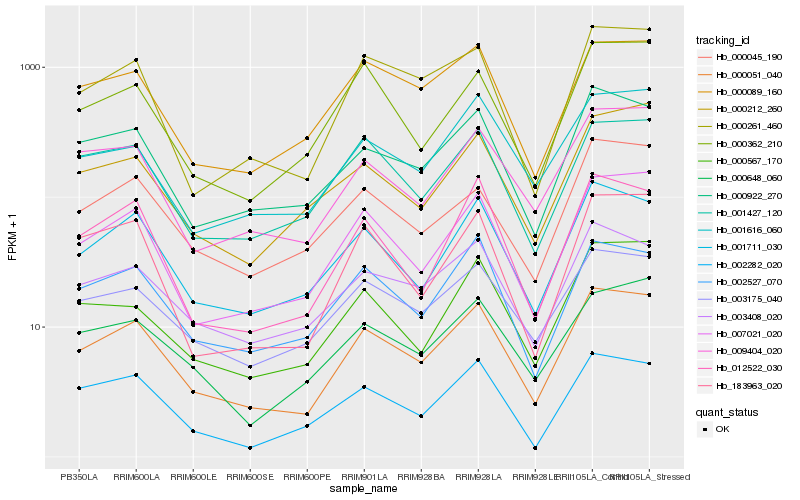
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000051_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC102597788 [Solanum tuberosum] |
| 2 |
Hb_007021_020 |
0.0689325583 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001711_030 |
0.0758318271 |
- |
- |
PREDICTED: uncharacterized protein LOC105642409 [Jatropha curcas] |
| 4 |
Hb_000922_270 |
0.0824675172 |
- |
- |
iron-sulfur cluster scaffold protein [Hevea brasiliensis] |
| 5 |
Hb_000261_460 |
0.087561869 |
- |
- |
60S ribosomal protein L13aA [Hevea brasiliensis] |
| 6 |
Hb_009404_020 |
0.0894587089 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003408_020 |
0.0908699352 |
- |
- |
hypothetical protein RCOM_0453040 [Ricinus communis] |
| 8 |
Hb_012522_030 |
0.092166935 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_003175_040 |
0.092194278 |
- |
- |
PREDICTED: katanin p60 ATPase-containing subunit A1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_183963_020 |
0.0936367524 |
- |
- |
PREDICTED: uncharacterized protein LOC105644615 [Jatropha curcas] |
| 11 |
Hb_002282_020 |
0.0953840217 |
- |
- |
PREDICTED: uncharacterized protein LOC105629828 [Jatropha curcas] |
| 12 |
Hb_002527_070 |
0.0955076192 |
- |
- |
axonemal dynein light chain, putative [Ricinus communis] |
| 13 |
Hb_000089_160 |
0.0955373635 |
- |
- |
60S ribosomal protein L5B [Hevea brasiliensis] |
| 14 |
Hb_000648_060 |
0.098256135 |
- |
- |
hypothetical protein POPTR_0019s10570g [Populus trichocarpa] |
| 15 |
Hb_001427_120 |
0.0986811588 |
- |
- |
PREDICTED: uncharacterized protein LOC105637787 [Jatropha curcas] |
| 16 |
Hb_000567_170 |
0.0988505832 |
- |
- |
PREDICTED: putative lipase YDR444W isoform X1 [Jatropha curcas] |
| 17 |
Hb_001616_060 |
0.0997265015 |
- |
- |
PREDICTED: DNA-directed RNA polymerase subunit 10-like protein [Jatropha curcas] |
| 18 |
Hb_000362_210 |
0.1010729428 |
- |
- |
PREDICTED: transcription elongation factor 1 homolog [Jatropha curcas] |
| 19 |
Hb_000045_190 |
0.1014803844 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit F [Jatropha curcas] |
| 20 |
Hb_000212_260 |
0.1034225295 |
- |
- |
acyl-carrier-protein [Triadica sebifera] |