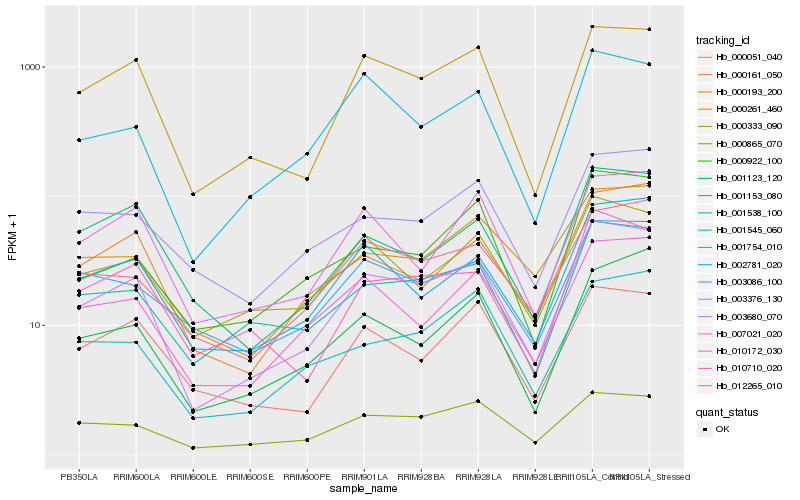
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003680_070 |
0.0 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000261_460 |
0.0906108694 |
- |
- |
60S ribosomal protein L13aA [Hevea brasiliensis] |
| 3 |
Hb_001538_100 |
0.1045923947 |
- |
- |
PREDICTED: histone deacetylase 6 [Jatropha curcas] |
| 4 |
Hb_010172_030 |
0.1055569722 |
- |
- |
hypothetical protein JCGZ_04668 [Jatropha curcas] |
| 5 |
Hb_000865_070 |
0.1061514915 |
- |
- |
Protein FAR1-RELATED SEQUENCE 3 [Glycine soja] |
| 6 |
Hb_002781_020 |
0.1078201421 |
- |
- |
PREDICTED: uncharacterized protein LOC105644398 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001754_010 |
0.1108031841 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 8 |
Hb_010710_020 |
0.1120267236 |
- |
- |
PREDICTED: uncharacterized protein LOC103322096 [Prunus mume] |
| 9 |
Hb_000161_050 |
0.1121200272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_012265_010 |
0.1127531897 |
- |
- |
hypothetical protein CISIN_1g026883mg [Citrus sinensis] |
| 11 |
Hb_000333_090 |
0.115487535 |
- |
- |
PREDICTED: uncharacterized protein LOC102617374 isoform X1 [Citrus sinensis] |
| 12 |
Hb_000922_100 |
0.1176048587 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |
| 13 |
Hb_007021_020 |
0.1176186286 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000193_200 |
0.1185371338 |
- |
- |
rotamase, putative [Ricinus communis] |
| 15 |
Hb_001123_120 |
0.1189183889 |
- |
- |
Uncharacterized protein TCM_042776 [Theobroma cacao] |
| 16 |
Hb_001545_060 |
0.119774285 |
- |
- |
PREDICTED: uncharacterized protein LOC105643880 [Jatropha curcas] |
| 17 |
Hb_003376_130 |
0.1200835159 |
transcription factor |
TF Family: S1Fa-like |
DNA-binding protein S1FA, putative [Ricinus communis] |
| 18 |
Hb_003086_100 |
0.1209816432 |
- |
- |
hypothetical protein JCGZ_08729 [Jatropha curcas] |
| 19 |
Hb_000051_040 |
0.12187806 |
- |
- |
PREDICTED: uncharacterized protein LOC102597788 [Solanum tuberosum] |
| 20 |
Hb_001153_080 |
0.1224012408 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL19 [Jatropha curcas] |