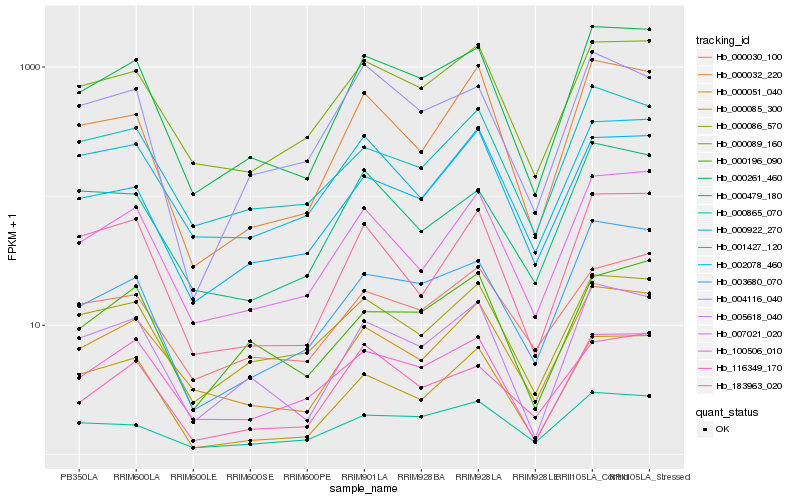
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_460 |
0.0 |
- |
- |
60S ribosomal protein L13aA [Hevea brasiliensis] |
| 2 |
Hb_005618_040 |
0.0736244363 |
- |
- |
- |
| 3 |
Hb_000089_160 |
0.0851148563 |
- |
- |
60S ribosomal protein L5B [Hevea brasiliensis] |
| 4 |
Hb_000051_040 |
0.087561869 |
- |
- |
PREDICTED: uncharacterized protein LOC102597788 [Solanum tuberosum] |
| 5 |
Hb_003680_070 |
0.0906108694 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_007021_020 |
0.0948636345 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_100506_010 |
0.1017166055 |
- |
- |
PREDICTED: uncharacterized protein LOC105133651 isoform X2 [Populus euphratica] |
| 8 |
Hb_000865_070 |
0.1029025118 |
- |
- |
Protein FAR1-RELATED SEQUENCE 3 [Glycine soja] |
| 9 |
Hb_000086_570 |
0.1042306089 |
- |
- |
hypothetical protein JCGZ_24934 [Jatropha curcas] |
| 10 |
Hb_116349_170 |
0.1061449044 |
- |
- |
PREDICTED: uncharacterized protein LOC105649815 [Jatropha curcas] |
| 11 |
Hb_000196_090 |
0.1077187596 |
- |
- |
axonemal dynein light chain, putative [Ricinus communis] |
| 12 |
Hb_000085_300 |
0.1079245772 |
- |
- |
- |
| 13 |
Hb_000032_220 |
0.1098112862 |
- |
- |
PREDICTED: abscisic acid receptor PYL8-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000030_100 |
0.1104114898 |
- |
- |
PREDICTED: short-chain type dehydrogenase/reductase isoform X2 [Jatropha curcas] |
| 15 |
Hb_183963_020 |
0.1107917667 |
- |
- |
PREDICTED: uncharacterized protein LOC105644615 [Jatropha curcas] |
| 16 |
Hb_000922_270 |
0.1128089384 |
- |
- |
iron-sulfur cluster scaffold protein [Hevea brasiliensis] |
| 17 |
Hb_002078_460 |
0.1142768654 |
- |
- |
- |
| 18 |
Hb_001427_120 |
0.1148114608 |
- |
- |
PREDICTED: uncharacterized protein LOC105637787 [Jatropha curcas] |
| 19 |
Hb_000479_180 |
0.1158130969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004116_040 |
0.1172402539 |
transcription factor |
TF Family: HMG |
hypothetical protein POPTR_0010s19720g [Populus trichocarpa] |