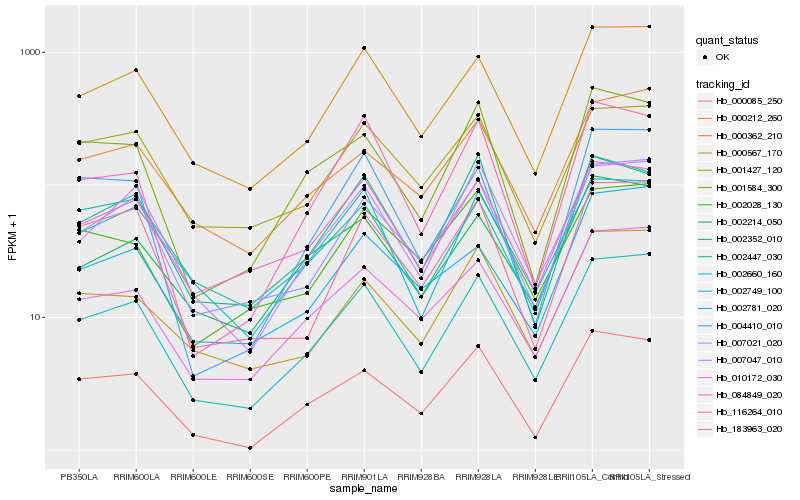
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002660_160 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_000362_210 |
0.0606710177 |
- |
- |
PREDICTED: transcription elongation factor 1 homolog [Jatropha curcas] |
| 3 |
Hb_007021_020 |
0.0685129585 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_010172_030 |
0.075600118 |
- |
- |
hypothetical protein JCGZ_04668 [Jatropha curcas] |
| 5 |
Hb_007047_010 |
0.0826479899 |
- |
- |
PREDICTED: expansin-A13 [Jatropha curcas] |
| 6 |
Hb_000085_250 |
0.0912099178 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 7 |
Hb_002749_100 |
0.0917416523 |
- |
- |
PREDICTED: ubiquitin domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_000212_260 |
0.0920620358 |
- |
- |
acyl-carrier-protein [Triadica sebifera] |
| 9 |
Hb_000567_170 |
0.0930928375 |
- |
- |
PREDICTED: putative lipase YDR444W isoform X1 [Jatropha curcas] |
| 10 |
Hb_084849_020 |
0.0950184777 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 11 |
Hb_002214_050 |
0.099035148 |
- |
- |
PREDICTED: probable CDP-diacylglycerol--inositol 3-phosphatidyltransferase 2 [Jatropha curcas] |
| 12 |
Hb_004410_010 |
0.1002240213 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 13 |
Hb_183963_020 |
0.1003551526 |
- |
- |
PREDICTED: uncharacterized protein LOC105644615 [Jatropha curcas] |
| 14 |
Hb_001584_300 |
0.1024339658 |
- |
- |
PREDICTED: pyroglutamyl-peptidase 1-like protein [Jatropha curcas] |
| 15 |
Hb_001427_120 |
0.107673449 |
- |
- |
PREDICTED: uncharacterized protein LOC105637787 [Jatropha curcas] |
| 16 |
Hb_002781_020 |
0.1083153779 |
- |
- |
PREDICTED: uncharacterized protein LOC105644398 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002028_130 |
0.1083789228 |
- |
- |
PREDICTED: MHC class II regulatory factor RFX1-like [Jatropha curcas] |
| 18 |
Hb_002352_010 |
0.1092350917 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 19 |
Hb_116264_010 |
0.1097389058 |
- |
- |
hypothetical protein JCGZ_05585 [Jatropha curcas] |
| 20 |
Hb_002447_030 |
0.1107394137 |
- |
- |
PREDICTED: cytochrome b5 domain-containing protein RLF [Jatropha curcas] |