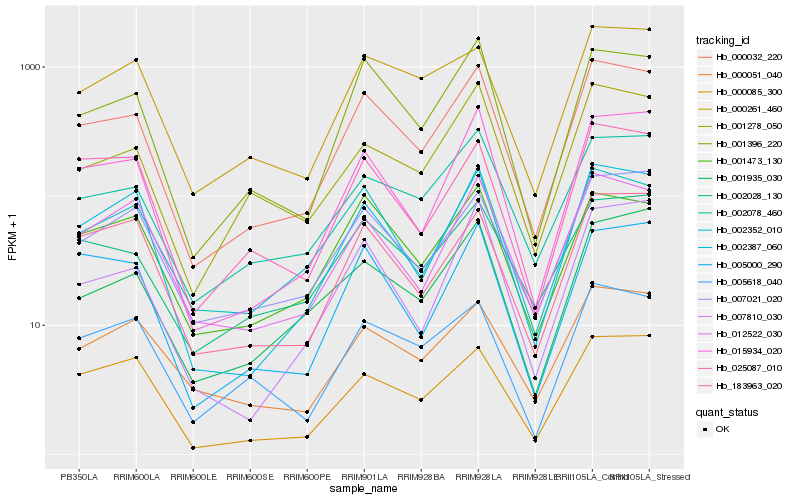
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000032_220 |
0.0 |
- |
- |
PREDICTED: abscisic acid receptor PYL8-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_001396_220 |
0.0720795024 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 3 |
Hb_015934_020 |
0.0804588938 |
- |
- |
PREDICTED: uncharacterized protein LOC105638669 [Jatropha curcas] |
| 4 |
Hb_002387_060 |
0.0857128152 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002078_460 |
0.0888263596 |
- |
- |
- |
| 6 |
Hb_183963_020 |
0.0985282606 |
- |
- |
PREDICTED: uncharacterized protein LOC105644615 [Jatropha curcas] |
| 7 |
Hb_002352_010 |
0.0986983996 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 8 |
Hb_007021_020 |
0.0996279544 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000085_300 |
0.1003599668 |
- |
- |
- |
| 10 |
Hb_005000_290 |
0.1005038745 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_07430 [Jatropha curcas] |
| 11 |
Hb_012522_030 |
0.1008027863 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_002028_130 |
0.1010631544 |
- |
- |
PREDICTED: MHC class II regulatory factor RFX1-like [Jatropha curcas] |
| 13 |
Hb_001473_130 |
0.1032429169 |
- |
- |
PREDICTED: ubiquinone biosynthesis monooxygenase COQ6 [Jatropha curcas] |
| 14 |
Hb_001278_050 |
0.1041397778 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 15 |
Hb_025087_010 |
0.104477369 |
- |
- |
PREDICTED: RING-H2 finger protein ATL56 [Jatropha curcas] |
| 16 |
Hb_000051_040 |
0.1063954068 |
- |
- |
PREDICTED: uncharacterized protein LOC102597788 [Solanum tuberosum] |
| 17 |
Hb_005618_040 |
0.1085226255 |
- |
- |
- |
| 18 |
Hb_007810_030 |
0.1090154215 |
- |
- |
PREDICTED: uncharacterized protein LOC105647693 [Jatropha curcas] |
| 19 |
Hb_000261_460 |
0.1098112862 |
- |
- |
60S ribosomal protein L13aA [Hevea brasiliensis] |
| 20 |
Hb_001935_030 |
0.1102766387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |