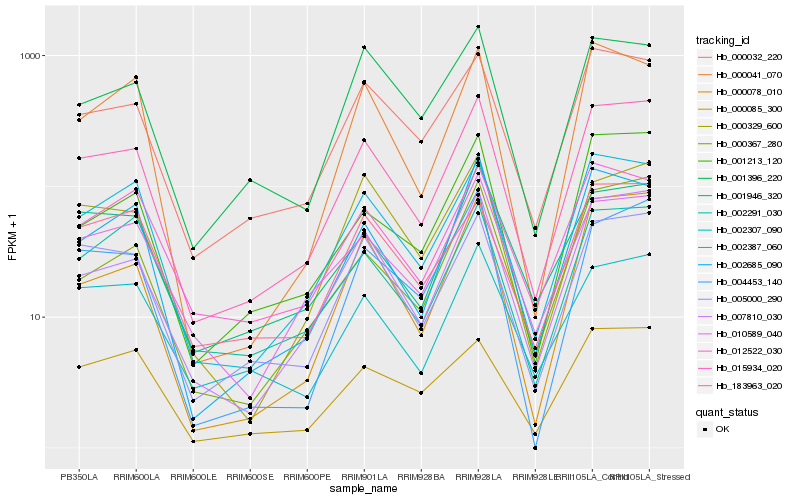
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015934_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638669 [Jatropha curcas] |
| 2 |
Hb_007810_030 |
0.0720719594 |
- |
- |
PREDICTED: uncharacterized protein LOC105647693 [Jatropha curcas] |
| 3 |
Hb_005000_290 |
0.0772733959 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_07430 [Jatropha curcas] |
| 4 |
Hb_002387_060 |
0.0773052707 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000032_220 |
0.0804588938 |
- |
- |
PREDICTED: abscisic acid receptor PYL8-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000078_010 |
0.0971870814 |
- |
- |
hypothetical protein POPTR_0006s06510g, partial [Populus trichocarpa] |
| 7 |
Hb_002685_090 |
0.0989043672 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0004s04590g [Populus trichocarpa] |
| 8 |
Hb_010589_040 |
0.1039622154 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000367_280 |
0.1042908076 |
- |
- |
PREDICTED: flavin-containing monooxygenase FMO GS-OX-like 9 [Jatropha curcas] |
| 10 |
Hb_001213_120 |
0.1042977274 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Brassica rapa] |
| 11 |
Hb_000085_300 |
0.1060294069 |
- |
- |
- |
| 12 |
Hb_000329_600 |
0.1062744021 |
- |
- |
PREDICTED: cell number regulator 6-like isoform X2 [Populus euphratica] |
| 13 |
Hb_001396_220 |
0.1086390321 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 14 |
Hb_183963_020 |
0.1088022033 |
- |
- |
PREDICTED: uncharacterized protein LOC105644615 [Jatropha curcas] |
| 15 |
Hb_002307_090 |
0.1095601205 |
- |
- |
magnesium transporter CorA-like family protein [Populus trichocarpa] |
| 16 |
Hb_004453_140 |
0.1101882561 |
- |
- |
PREDICTED: uncharacterized protein LOC104880181 [Vitis vinifera] |
| 17 |
Hb_012522_030 |
0.1103895301 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_001946_320 |
0.1115809224 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g55270-like [Jatropha curcas] |
| 19 |
Hb_002291_030 |
0.114008547 |
- |
- |
PREDICTED: vesicle transport protein SFT2B [Jatropha curcas] |
| 20 |
Hb_000041_070 |
0.114979971 |
- |
- |
hypothetical protein POPTR_0001s34870g [Populus trichocarpa] |