| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
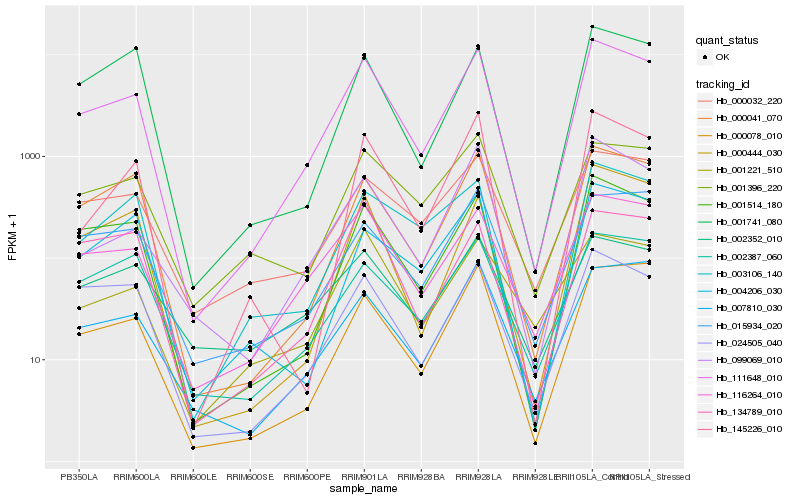
Hb_111648_010 |
0.0 |
rubber biosynthesis |
Gene Name: REF1 |
rubber elongation factor [Hevea brasiliensis] |
| 2 |
Hb_001514_180 |
0.0921270912 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_16067 [Jatropha curcas] |
| 3 |
Hb_116264_010 |
0.0965646044 |
- |
- |
hypothetical protein JCGZ_05585 [Jatropha curcas] |
| 4 |
Hb_000041_070 |
0.1130948421 |
- |
- |
hypothetical protein POPTR_0001s34870g [Populus trichocarpa] |
| 5 |
Hb_000078_010 |
0.1196517404 |
- |
- |
hypothetical protein POPTR_0006s06510g, partial [Populus trichocarpa] |
| 6 |
Hb_024505_040 |
0.1210279068 |
- |
- |
homogentisate geranylgeranyl transferase [Hevea brasiliensis] |
| 7 |
Hb_000444_030 |
0.1261120092 |
- |
- |
PREDICTED: expansin-A20 [Jatropha curcas] |
| 8 |
Hb_145226_010 |
0.1262559238 |
- |
- |
PREDICTED: uncharacterized protein LOC100262861 [Vitis vinifera] |
| 9 |
Hb_000032_220 |
0.130310572 |
- |
- |
PREDICTED: abscisic acid receptor PYL8-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_007810_030 |
0.1309378194 |
- |
- |
PREDICTED: uncharacterized protein LOC105647693 [Jatropha curcas] |
| 11 |
Hb_099069_010 |
0.1310631102 |
- |
- |
S-adenosyl-methionine-sterol-C- methyltransferase [Ricinus communis] |
| 12 |
Hb_001396_220 |
0.1311313099 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 13 |
Hb_003106_140 |
0.135624357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001221_510 |
0.136150992 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 15 |
Hb_002387_060 |
0.1393141042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_015934_020 |
0.1397975616 |
- |
- |
PREDICTED: uncharacterized protein LOC105638669 [Jatropha curcas] |
| 17 |
Hb_001741_080 |
0.1413412582 |
rubber biosynthesis |
Gene Name: REF8 |
small rubber particle protein [Hevea brasiliensis] |
| 18 |
Hb_002352_010 |
0.1494612632 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 19 |
Hb_004206_030 |
0.1495239767 |
- |
- |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 20 |
Hb_134789_010 |
0.1513583115 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |