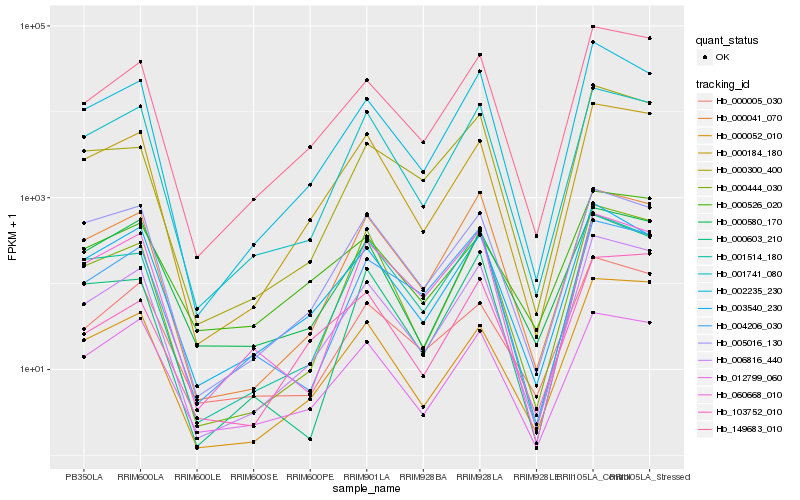
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006816_440 |
0.0 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 2 |
Hb_149683_010 |
0.0413547035 |
rubber biosynthesis |
Gene Name: REF2 |
RecName: Full=Rubber elongation factor protein; Short=REF; AltName: Allergen=Hev b 1 [Hevea brasiliensis] |
| 3 |
Hb_002235_230 |
0.0733539238 |
- |
- |
latex abundant protein 1 [Hevea brasiliensis] |
| 4 |
Hb_000184_180 |
0.081145959 |
- |
- |
putative C3HC4-type RING zinc finger protein [Hevea brasiliensis] |
| 5 |
Hb_000444_030 |
0.1004224991 |
- |
- |
PREDICTED: expansin-A20 [Jatropha curcas] |
| 6 |
Hb_001741_080 |
0.1012493004 |
rubber biosynthesis |
Gene Name: REF8 |
small rubber particle protein [Hevea brasiliensis] |
| 7 |
Hb_000052_010 |
0.1059068743 |
- |
- |
hypothetical protein M569_08416, partial [Genlisea aurea] |
| 8 |
Hb_003540_230 |
0.1062632434 |
- |
- |
PREDICTED: sphingosine kinase 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000005_030 |
0.108703839 |
- |
- |
hypothetical protein POPTR_0009s00870g [Populus trichocarpa] |
| 10 |
Hb_000300_400 |
0.1161937568 |
- |
- |
hypothetical protein PHAVU_001G114100g [Phaseolus vulgaris] |
| 11 |
Hb_060668_010 |
0.1188331712 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 12 |
Hb_000041_070 |
0.1195456203 |
- |
- |
hypothetical protein POPTR_0001s34870g [Populus trichocarpa] |
| 13 |
Hb_004206_030 |
0.1196159843 |
- |
- |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 14 |
Hb_012799_060 |
0.1223884065 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 15 |
Hb_000603_210 |
0.1230699823 |
- |
- |
hypothetical protein POPTR_0006s23660g [Populus trichocarpa] |
| 16 |
Hb_000580_170 |
0.123848452 |
- |
- |
PREDICTED: arginase 1, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000526_020 |
0.124530537 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 18 |
Hb_001514_180 |
0.1290341812 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_16067 [Jatropha curcas] |
| 19 |
Hb_103752_010 |
0.1300724425 |
- |
- |
PREDICTED: uncharacterized protein At5g39865-like [Jatropha curcas] |
| 20 |
Hb_005016_130 |
0.1306468074 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |