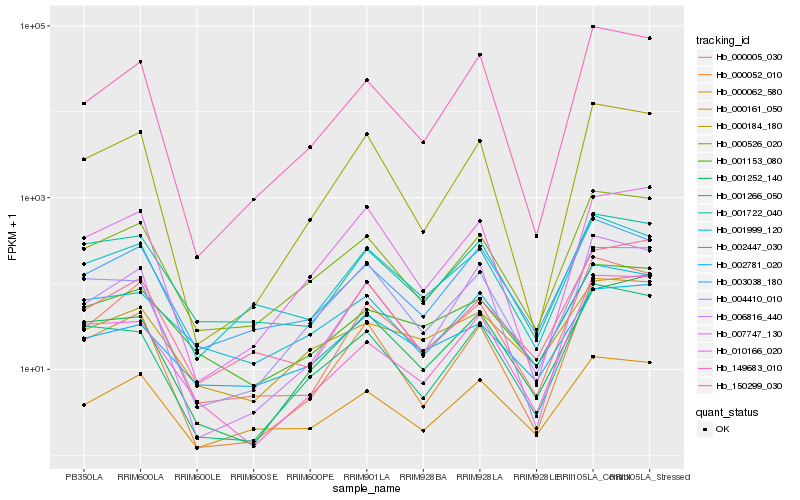
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000526_020 |
0.0 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 2 |
Hb_000062_580 |
0.0933346792 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_001266_050 |
0.1008567999 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like isoform X2 [Elaeis guineensis] |
| 4 |
Hb_000052_010 |
0.1017541951 |
- |
- |
hypothetical protein M569_08416, partial [Genlisea aurea] |
| 5 |
Hb_000005_030 |
0.1027446503 |
- |
- |
hypothetical protein POPTR_0009s00870g [Populus trichocarpa] |
| 6 |
Hb_003038_180 |
0.1102936311 |
- |
- |
PREDICTED: uncharacterized protein LOC105649131 [Jatropha curcas] |
| 7 |
Hb_000184_180 |
0.112478985 |
- |
- |
putative C3HC4-type RING zinc finger protein [Hevea brasiliensis] |
| 8 |
Hb_150299_030 |
0.117802169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001153_080 |
0.1178690235 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL19 [Jatropha curcas] |
| 10 |
Hb_149683_010 |
0.1191832323 |
rubber biosynthesis |
Gene Name: REF2 |
RecName: Full=Rubber elongation factor protein; Short=REF; AltName: Allergen=Hev b 1 [Hevea brasiliensis] |
| 11 |
Hb_001722_040 |
0.1219326232 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 variant 1A-like [Jatropha curcas] |
| 12 |
Hb_000161_050 |
0.1225803007 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_010166_020 |
0.1239067427 |
transcription factor |
TF Family: NF-YC |
ccaat-binding transcription factor, putative [Ricinus communis] |
| 14 |
Hb_006816_440 |
0.124530537 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 15 |
Hb_002781_020 |
0.1264042726 |
- |
- |
PREDICTED: uncharacterized protein LOC105644398 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001252_140 |
0.1275101747 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_001999_120 |
0.1277566197 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 18 |
Hb_002447_030 |
0.1280564407 |
- |
- |
PREDICTED: cytochrome b5 domain-containing protein RLF [Jatropha curcas] |
| 19 |
Hb_004410_010 |
0.1331525748 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 20 |
Hb_007747_130 |
0.1354485288 |
- |
- |
PREDICTED: uncharacterized protein LOC105635879 [Jatropha curcas] |