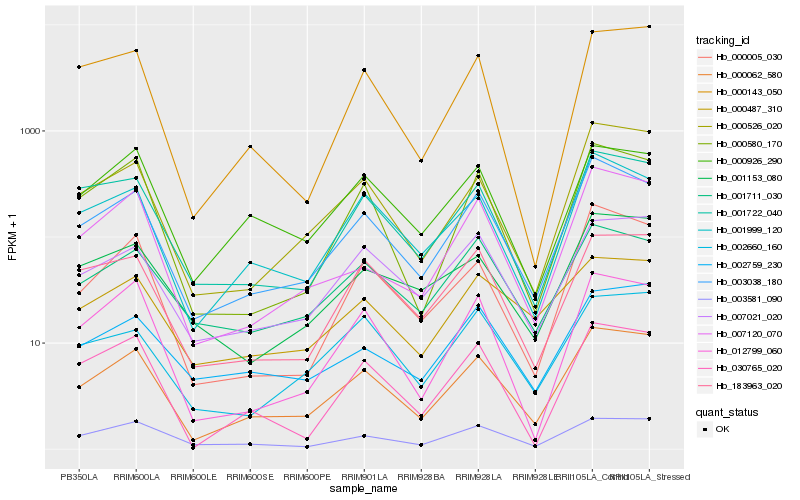
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_580 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_003038_180 |
0.0714327773 |
- |
- |
PREDICTED: uncharacterized protein LOC105649131 [Jatropha curcas] |
| 3 |
Hb_000526_020 |
0.0933346792 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 4 |
Hb_001999_120 |
0.0969998482 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 5 |
Hb_007021_020 |
0.1021401337 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003581_090 |
0.1043364171 |
- |
- |
PREDICTED: galactose oxidase [Jatropha curcas] |
| 7 |
Hb_001722_040 |
0.1087191696 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 variant 1A-like [Jatropha curcas] |
| 8 |
Hb_000005_030 |
0.1138560323 |
- |
- |
hypothetical protein POPTR_0009s00870g [Populus trichocarpa] |
| 9 |
Hb_000580_170 |
0.1139505168 |
- |
- |
PREDICTED: arginase 1, mitochondrial [Jatropha curcas] |
| 10 |
Hb_183963_020 |
0.1142013066 |
- |
- |
PREDICTED: uncharacterized protein LOC105644615 [Jatropha curcas] |
| 11 |
Hb_000926_290 |
0.1143962802 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_012799_060 |
0.1186489701 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 13 |
Hb_007120_070 |
0.1219506457 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 14 |
Hb_030765_020 |
0.1234176561 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_000143_050 |
0.123591384 |
- |
- |
PREDICTED: MFP1 attachment factor 1-like [Jatropha curcas] |
| 16 |
Hb_002759_230 |
0.1236460778 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 25 [Jatropha curcas] |
| 17 |
Hb_000487_310 |
0.1240851225 |
- |
- |
PREDICTED: uncharacterized protein LOC105646211 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001711_030 |
0.1245011773 |
- |
- |
PREDICTED: uncharacterized protein LOC105642409 [Jatropha curcas] |
| 19 |
Hb_001153_080 |
0.1246971758 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL19 [Jatropha curcas] |
| 20 |
Hb_002660_160 |
0.1248789947 |
- |
- |
protein binding protein, putative [Ricinus communis] |