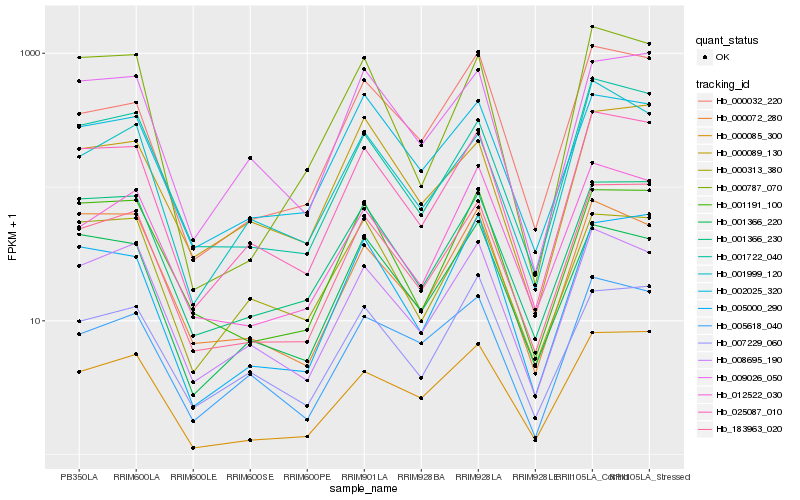
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025087_010 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL56 [Jatropha curcas] |
| 2 |
Hb_183963_020 |
0.0581526738 |
- |
- |
PREDICTED: uncharacterized protein LOC105644615 [Jatropha curcas] |
| 3 |
Hb_001366_230 |
0.06959112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_008695_190 |
0.0737896815 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 17 [Jatropha curcas] |
| 5 |
Hb_009026_050 |
0.0754854236 |
- |
- |
vacuolar ATPase F subunit [Hevea brasiliensis] |
| 6 |
Hb_001722_040 |
0.0774459327 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 variant 1A-like [Jatropha curcas] |
| 7 |
Hb_005000_290 |
0.0856729214 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_07430 [Jatropha curcas] |
| 8 |
Hb_000085_300 |
0.0890054347 |
- |
- |
- |
| 9 |
Hb_001366_220 |
0.089740501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007229_060 |
0.0941245061 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g44880 [Jatropha curcas] |
| 11 |
Hb_001191_100 |
0.0945990146 |
- |
- |
F-actin capping protein beta subunit [Populus trichocarpa] |
| 12 |
Hb_005618_040 |
0.0954759329 |
- |
- |
- |
| 13 |
Hb_001999_120 |
0.0993215532 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 14 |
Hb_000313_380 |
0.1018121965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000089_130 |
0.1042573929 |
- |
- |
mitochondrial ribosomal protein S16, putative [Ricinus communis] |
| 16 |
Hb_000032_220 |
0.104477369 |
- |
- |
PREDICTED: abscisic acid receptor PYL8-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_002025_320 |
0.1049583165 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit I-like [Jatropha curcas] |
| 18 |
Hb_000072_280 |
0.1051554856 |
- |
- |
PREDICTED: biogenesis of lysosome-related organelles complex 1 subunit 2-like [Jatropha curcas] |
| 19 |
Hb_000787_070 |
0.1064836834 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 20 |
Hb_012522_030 |
0.1066522367 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |