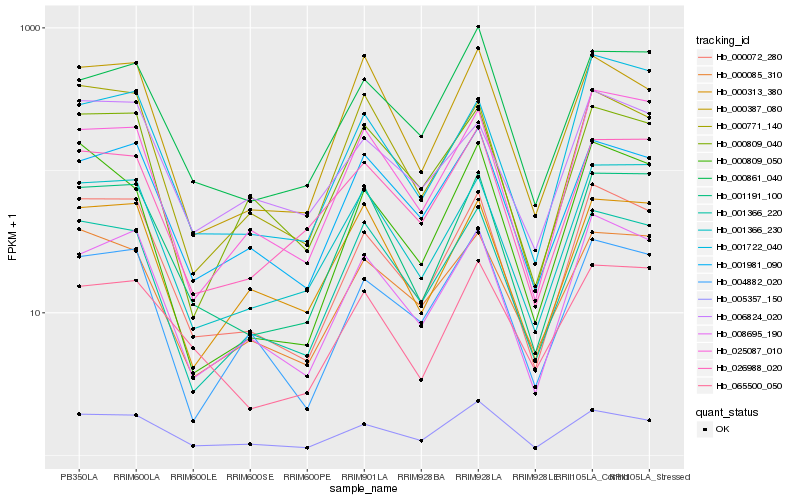
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000072_280 |
0.0 |
- |
- |
PREDICTED: biogenesis of lysosome-related organelles complex 1 subunit 2-like [Jatropha curcas] |
| 2 |
Hb_008695_190 |
0.0727014757 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 17 [Jatropha curcas] |
| 3 |
Hb_001191_100 |
0.0935686309 |
- |
- |
F-actin capping protein beta subunit [Populus trichocarpa] |
| 4 |
Hb_004882_020 |
0.095080328 |
- |
- |
- |
| 5 |
Hb_001366_220 |
0.09605277 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000085_310 |
0.0975512162 |
- |
- |
hypothetical protein JCGZ_00974 [Jatropha curcas] |
| 7 |
Hb_001366_230 |
0.0979316856 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005357_150 |
0.101085236 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_001981_090 |
0.1011626015 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 1-like [Prunus mume] |
| 10 |
Hb_000809_040 |
0.1025056512 |
- |
- |
Serine/threonine-protein kinase SAPK10, putative [Ricinus communis] |
| 11 |
Hb_025087_010 |
0.1051554856 |
- |
- |
PREDICTED: RING-H2 finger protein ATL56 [Jatropha curcas] |
| 12 |
Hb_006824_020 |
0.1072497787 |
- |
- |
PREDICTED: thioredoxin domain-containing protein 9 homolog [Jatropha curcas] |
| 13 |
Hb_000771_140 |
0.10868193 |
- |
- |
PREDICTED: abscisic acid receptor PYL8 [Jatropha curcas] |
| 14 |
Hb_000809_050 |
0.1125239448 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639663 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000387_080 |
0.1151948909 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000313_380 |
0.1172861403 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_026988_020 |
0.1180260496 |
- |
- |
outward rectifying K+ channel [Hevea brasiliensis] |
| 18 |
Hb_001722_040 |
0.1192411676 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 variant 1A-like [Jatropha curcas] |
| 19 |
Hb_000861_040 |
0.1203530485 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 1 [Jatropha curcas] |
| 20 |
Hb_065500_050 |
0.121847966 |
- |
- |
lipoate-protein ligase B, putative [Ricinus communis] |