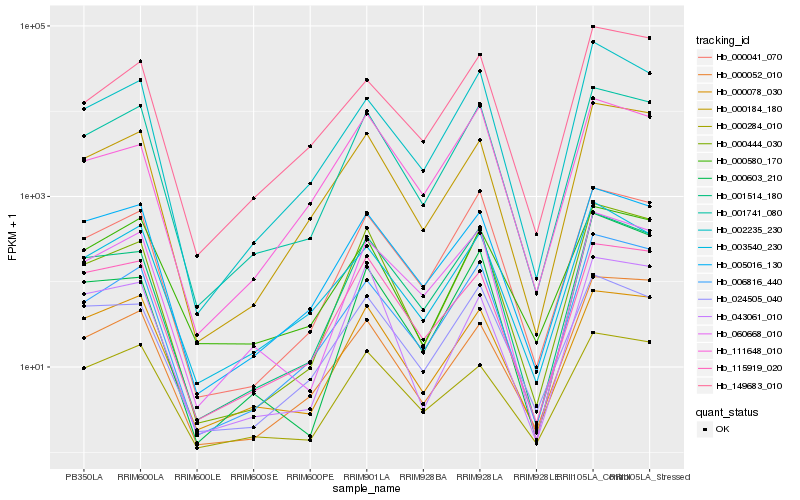
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000444_030 |
0.0 |
- |
- |
PREDICTED: expansin-A20 [Jatropha curcas] |
| 2 |
Hb_001741_080 |
0.0868264023 |
rubber biosynthesis |
Gene Name: REF8 |
small rubber particle protein [Hevea brasiliensis] |
| 3 |
Hb_000184_180 |
0.0898334992 |
- |
- |
putative C3HC4-type RING zinc finger protein [Hevea brasiliensis] |
| 4 |
Hb_001514_180 |
0.094060761 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_16067 [Jatropha curcas] |
| 5 |
Hb_006816_440 |
0.1004224991 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 6 |
Hb_000041_070 |
0.1167952256 |
- |
- |
hypothetical protein POPTR_0001s34870g [Populus trichocarpa] |
| 7 |
Hb_043061_010 |
0.121950068 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 8 |
Hb_060668_010 |
0.1230467952 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 9 |
Hb_000603_210 |
0.1257342999 |
- |
- |
hypothetical protein POPTR_0006s23660g [Populus trichocarpa] |
| 10 |
Hb_002235_230 |
0.1259888953 |
- |
- |
latex abundant protein 1 [Hevea brasiliensis] |
| 11 |
Hb_111648_010 |
0.1261120092 |
rubber biosynthesis |
Gene Name: REF1 |
rubber elongation factor [Hevea brasiliensis] |
| 12 |
Hb_115919_020 |
0.1284184934 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 13 |
Hb_005016_130 |
0.1305583349 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 14 |
Hb_149683_010 |
0.132052606 |
rubber biosynthesis |
Gene Name: REF2 |
RecName: Full=Rubber elongation factor protein; Short=REF; AltName: Allergen=Hev b 1 [Hevea brasiliensis] |
| 15 |
Hb_024505_040 |
0.1379129326 |
- |
- |
homogentisate geranylgeranyl transferase [Hevea brasiliensis] |
| 16 |
Hb_000284_010 |
0.1394180316 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 17 |
Hb_000052_010 |
0.1395337444 |
- |
- |
hypothetical protein M569_08416, partial [Genlisea aurea] |
| 18 |
Hb_000580_170 |
0.1445196245 |
- |
- |
PREDICTED: arginase 1, mitochondrial [Jatropha curcas] |
| 19 |
Hb_003540_230 |
0.1465584647 |
- |
- |
PREDICTED: sphingosine kinase 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000078_030 |
0.1485504218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |