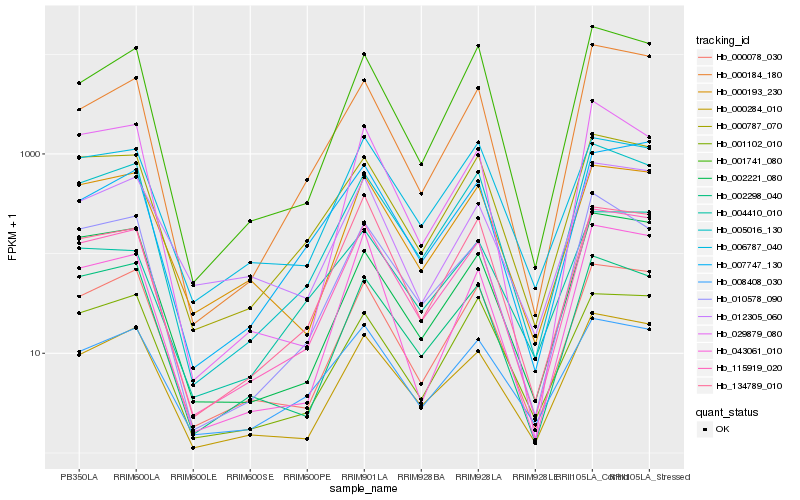
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_115919_020 |
0.0 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 2 |
Hb_000284_010 |
0.0603515719 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 3 |
Hb_005016_130 |
0.0682681327 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 4 |
Hb_000078_030 |
0.071731751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000787_070 |
0.079505554 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 6 |
Hb_000193_230 |
0.0888979803 |
- |
- |
PREDICTED: NEP1-interacting protein-like 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002298_040 |
0.0959840203 |
- |
- |
PREDICTED: uncharacterized protein LOC105633642 [Jatropha curcas] |
| 8 |
Hb_010578_090 |
0.0979346387 |
- |
- |
HEV2.1 [Hevea brasiliensis] |
| 9 |
Hb_043061_010 |
0.0980456936 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 10 |
Hb_001741_080 |
0.0995329742 |
rubber biosynthesis |
Gene Name: REF8 |
small rubber particle protein [Hevea brasiliensis] |
| 11 |
Hb_002221_080 |
0.1005082512 |
- |
- |
PREDICTED: uncharacterized protein LOC105639992 [Jatropha curcas] |
| 12 |
Hb_008408_030 |
0.1019325466 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 13 |
Hb_000184_180 |
0.1027113175 |
- |
- |
putative C3HC4-type RING zinc finger protein [Hevea brasiliensis] |
| 14 |
Hb_012305_060 |
0.1041439498 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 15 |
Hb_007747_130 |
0.1065100167 |
- |
- |
PREDICTED: uncharacterized protein LOC105635879 [Jatropha curcas] |
| 16 |
Hb_006787_040 |
0.1127371233 |
- |
- |
polyadenylate-binding protein, putative [Ricinus communis] |
| 17 |
Hb_004410_010 |
0.1127466805 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 18 |
Hb_134789_010 |
0.1131440047 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 19 |
Hb_001102_010 |
0.1149952732 |
- |
- |
PREDICTED: solanesyl diphosphate synthase 3, chloroplastic/mitochondrial-like isoform X2 [Glycine max] |
| 20 |
Hb_029879_080 |
0.1156142235 |
- |
- |
unknown [Populus trichocarpa] |