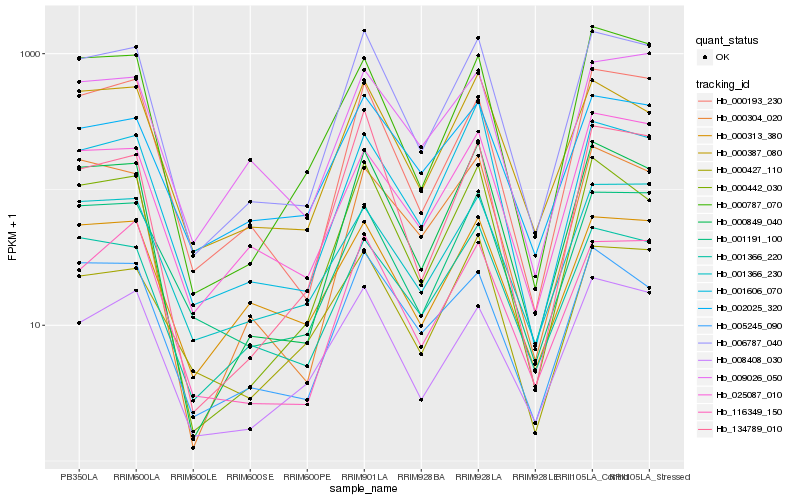
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006787_040 |
0.0 |
- |
- |
polyadenylate-binding protein, putative [Ricinus communis] |
| 2 |
Hb_000849_040 |
0.0632177967 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_000442_030 |
0.0829129052 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000193_230 |
0.083716671 |
- |
- |
PREDICTED: NEP1-interacting protein-like 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000387_080 |
0.088223849 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001366_220 |
0.0907226547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001191_100 |
0.0927077594 |
- |
- |
F-actin capping protein beta subunit [Populus trichocarpa] |
| 8 |
Hb_008408_030 |
0.0929518483 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 9 |
Hb_005245_090 |
0.0967821982 |
- |
- |
PREDICTED: peroxisomal membrane protein 13 [Jatropha curcas] |
| 10 |
Hb_001606_070 |
0.0986440645 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 11 |
Hb_002025_320 |
0.0990875444 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit I-like [Jatropha curcas] |
| 12 |
Hb_134789_010 |
0.1015070913 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 13 |
Hb_116349_150 |
0.1026816278 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 14 |
Hb_000787_070 |
0.1032894383 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 15 |
Hb_000304_020 |
0.105228026 |
- |
- |
beta-1,3-glucanase form RRII Gln 3 [Hevea brasiliensis] |
| 16 |
Hb_009026_050 |
0.1077594178 |
- |
- |
vacuolar ATPase F subunit [Hevea brasiliensis] |
| 17 |
Hb_025087_010 |
0.1079909397 |
- |
- |
PREDICTED: RING-H2 finger protein ATL56 [Jatropha curcas] |
| 18 |
Hb_000313_380 |
0.1081469899 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001366_230 |
0.1083151896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000427_110 |
0.1084272156 |
- |
- |
PREDICTED: DNA repair protein XRCC2 homolog isoform X2 [Jatropha curcas] |