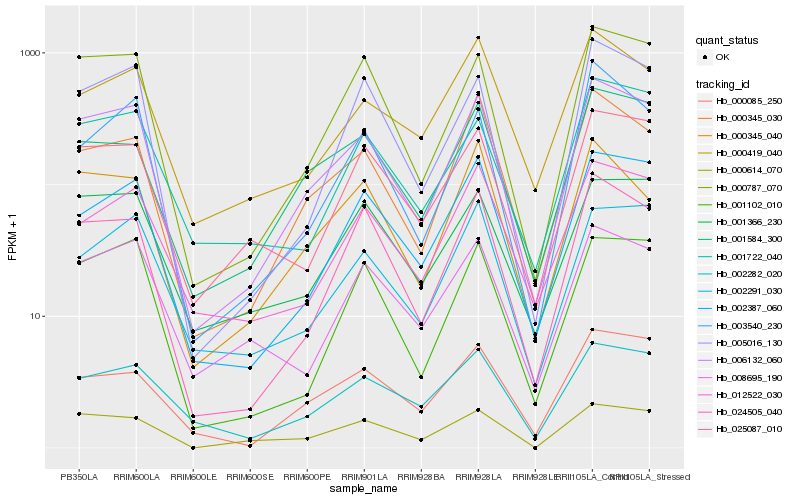
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006132_060 |
0.0 |
- |
- |
phospholipid/glycerol acyltransferase family protein [Populus trichocarpa] |
| 2 |
Hb_000787_070 |
0.0861750762 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 3 |
Hb_000345_030 |
0.0969554742 |
- |
- |
REF-like stress related protein 1 [Hevea brasiliensis] |
| 4 |
Hb_024505_040 |
0.1029893371 |
- |
- |
homogentisate geranylgeranyl transferase [Hevea brasiliensis] |
| 5 |
Hb_002387_060 |
0.1034728762 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001584_300 |
0.1082760767 |
- |
- |
PREDICTED: pyroglutamyl-peptidase 1-like protein [Jatropha curcas] |
| 7 |
Hb_005016_130 |
0.1095028593 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 8 |
Hb_000614_070 |
0.1111614596 |
- |
- |
- |
| 9 |
Hb_001102_010 |
0.113272769 |
- |
- |
PREDICTED: solanesyl diphosphate synthase 3, chloroplastic/mitochondrial-like isoform X2 [Glycine max] |
| 10 |
Hb_025087_010 |
0.1144800238 |
- |
- |
PREDICTED: RING-H2 finger protein ATL56 [Jatropha curcas] |
| 11 |
Hb_002282_020 |
0.1158280465 |
- |
- |
PREDICTED: uncharacterized protein LOC105629828 [Jatropha curcas] |
| 12 |
Hb_000345_040 |
0.1158925958 |
rubber biosynthesis |
Gene Name: REF4 |
- |
| 13 |
Hb_001366_230 |
0.1175084907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002291_030 |
0.1180322336 |
- |
- |
PREDICTED: vesicle transport protein SFT2B [Jatropha curcas] |
| 15 |
Hb_000085_250 |
0.1186675292 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 16 |
Hb_008695_190 |
0.1191329056 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 17 [Jatropha curcas] |
| 17 |
Hb_012522_030 |
0.1197351291 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_003540_230 |
0.1202402987 |
- |
- |
PREDICTED: sphingosine kinase 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001722_040 |
0.1210167215 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 variant 1A-like [Jatropha curcas] |
| 20 |
Hb_000419_040 |
0.1220325415 |
- |
- |
chorismate synthase [Hevea brasiliensis] |