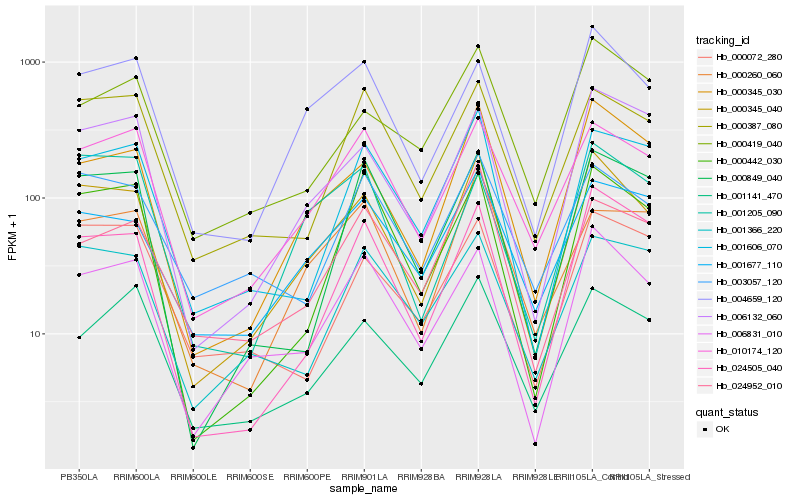
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000345_040 |
0.0 |
rubber biosynthesis |
Gene Name: REF4 |
- |
| 2 |
Hb_000345_030 |
0.0946239718 |
- |
- |
REF-like stress related protein 1 [Hevea brasiliensis] |
| 3 |
Hb_006831_010 |
0.1113793665 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_006132_060 |
0.1158925958 |
- |
- |
phospholipid/glycerol acyltransferase family protein [Populus trichocarpa] |
| 5 |
Hb_010174_120 |
0.1202330187 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1-like [Gossypium raimondii] |
| 6 |
Hb_004659_120 |
0.1207069079 |
rubber biosynthesis |
Gene Name: CPTL |
PREDICTED: dehydrodolichyl diphosphate syntase complex subunit NUS1 isoform X3 [Jatropha curcas] |
| 7 |
Hb_024505_040 |
0.1218947979 |
- |
- |
homogentisate geranylgeranyl transferase [Hevea brasiliensis] |
| 8 |
Hb_000387_080 |
0.1354588557 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001677_110 |
0.1362686218 |
- |
- |
Protein YIPF1, putative [Ricinus communis] |
| 10 |
Hb_001606_070 |
0.1374102006 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 11 |
Hb_000442_030 |
0.1385684083 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000419_040 |
0.1390229623 |
- |
- |
chorismate synthase [Hevea brasiliensis] |
| 13 |
Hb_003057_120 |
0.1407103758 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 14 |
Hb_000849_040 |
0.1437917195 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_001141_470 |
0.1451256156 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |
| 16 |
Hb_001366_220 |
0.1459619429 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001205_090 |
0.1497972573 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 18 |
Hb_000072_280 |
0.1499790745 |
- |
- |
PREDICTED: biogenesis of lysosome-related organelles complex 1 subunit 2-like [Jatropha curcas] |
| 19 |
Hb_024952_010 |
0.1506487413 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 20 |
Hb_000260_060 |
0.1508368205 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 6 [Jatropha curcas] |