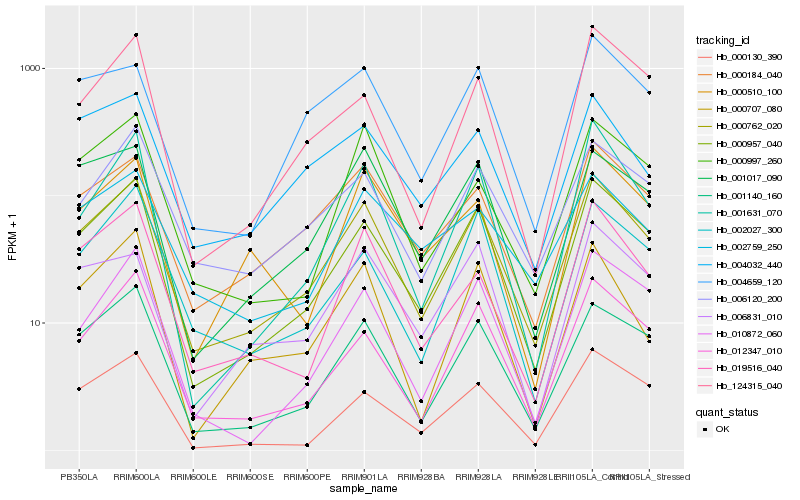
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000762_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 2 |
Hb_000957_040 |
0.0678174313 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 3 |
Hb_001140_160 |
0.0903548605 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 4 |
Hb_000184_040 |
0.0928647283 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
PREDICTED: acetyl-CoA acetyltransferase, cytosolic 1-like isoform X2 [Nelumbo nucifera] |
| 5 |
Hb_006120_200 |
0.1041049826 |
- |
- |
PREDICTED: uncharacterized protein LOC103333282 [Prunus mume] |
| 6 |
Hb_012347_010 |
0.1108264387 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |
| 7 |
Hb_001017_090 |
0.1167362498 |
- |
- |
- |
| 8 |
Hb_001631_070 |
0.1167851694 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 9 |
Hb_004659_120 |
0.1186585003 |
rubber biosynthesis |
Gene Name: CPTL |
PREDICTED: dehydrodolichyl diphosphate syntase complex subunit NUS1 isoform X3 [Jatropha curcas] |
| 10 |
Hb_019516_040 |
0.1207200161 |
desease resistance |
Gene Name: AAA |
Katanin p60 ATPase-containing subunit, putative [Ricinus communis] |
| 11 |
Hb_010872_060 |
0.121838851 |
transcription factor |
TF Family: MYB |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 12 |
Hb_004032_440 |
0.1227865878 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 1; Short=HMG-CoA reductase 1 [Hevea brasiliensis] |
| 13 |
Hb_006831_010 |
0.125893414 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_124315_040 |
0.1269468261 |
- |
- |
hypothetical protein 19 [Hevea brasiliensis] |
| 15 |
Hb_002027_300 |
0.1270016421 |
- |
- |
PREDICTED: rho GTPase-activating protein 2 [Jatropha curcas] |
| 16 |
Hb_002759_250 |
0.1282448849 |
rubber biosynthesis |
Gene Name: Phosphomevalonate kinase |
phosphomevalonate kinase [Hevea brasiliensis] |
| 17 |
Hb_000510_100 |
0.1294477426 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 18 |
Hb_000997_260 |
0.1296225531 |
- |
- |
PREDICTED: rRNA-processing protein EFG1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000707_080 |
0.1305087687 |
- |
- |
hypothetical protein AALP_AA4G156500, partial [Arabis alpina] |
| 20 |
Hb_000130_390 |
0.1312982351 |
- |
- |
PREDICTED: glutamate receptor 2.8-like [Pyrus x bretschneideri] |