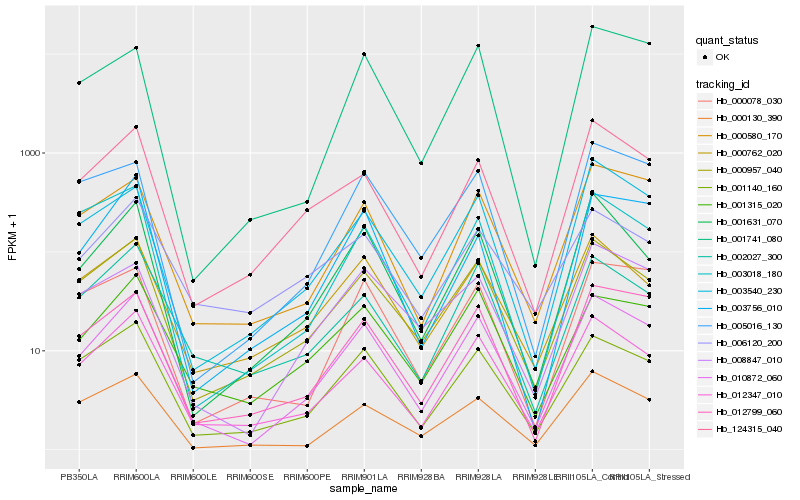
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010872_060 |
0.0 |
transcription factor |
TF Family: MYB |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 2 |
Hb_012347_010 |
0.0872591572 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |
| 3 |
Hb_000957_040 |
0.0949417196 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 4 |
Hb_001140_160 |
0.1081724875 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 5 |
Hb_001631_070 |
0.1081944446 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 6 |
Hb_124315_040 |
0.10939092 |
- |
- |
hypothetical protein 19 [Hevea brasiliensis] |
| 7 |
Hb_012799_060 |
0.1096180647 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 8 |
Hb_000130_390 |
0.1183389761 |
- |
- |
PREDICTED: glutamate receptor 2.8-like [Pyrus x bretschneideri] |
| 9 |
Hb_000580_170 |
0.1185514208 |
- |
- |
PREDICTED: arginase 1, mitochondrial [Jatropha curcas] |
| 10 |
Hb_002027_300 |
0.1206688009 |
- |
- |
PREDICTED: rho GTPase-activating protein 2 [Jatropha curcas] |
| 11 |
Hb_000762_020 |
0.121838851 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 12 |
Hb_001315_020 |
0.1237566012 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 13 |
Hb_003540_230 |
0.1271976549 |
- |
- |
PREDICTED: sphingosine kinase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_008847_010 |
0.1364756489 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 15 |
Hb_003756_010 |
0.1404431428 |
- |
- |
PREDICTED: uncharacterized protein LOC105642108 [Jatropha curcas] |
| 16 |
Hb_003018_180 |
0.1426703028 |
- |
- |
PREDICTED: WEB family protein At3g51220-like [Jatropha curcas] |
| 17 |
Hb_005016_130 |
0.1443621584 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 18 |
Hb_006120_200 |
0.1456751885 |
- |
- |
PREDICTED: uncharacterized protein LOC103333282 [Prunus mume] |
| 19 |
Hb_001741_080 |
0.1458830852 |
rubber biosynthesis |
Gene Name: REF8 |
small rubber particle protein [Hevea brasiliensis] |
| 20 |
Hb_000078_030 |
0.1465993744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |