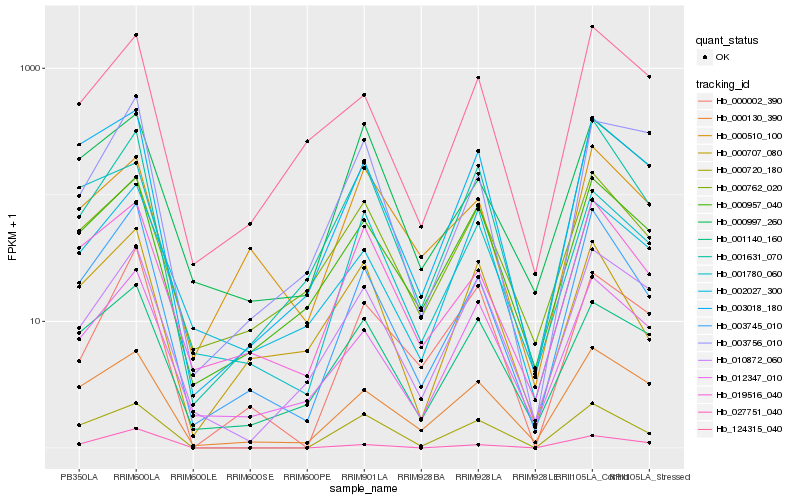
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003745_010 |
0.0 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |
| 2 |
Hb_001631_070 |
0.1315322306 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 3 |
Hb_019516_040 |
0.1329308766 |
desease resistance |
Gene Name: AAA |
Katanin p60 ATPase-containing subunit, putative [Ricinus communis] |
| 4 |
Hb_000130_390 |
0.1384991786 |
- |
- |
PREDICTED: glutamate receptor 2.8-like [Pyrus x bretschneideri] |
| 5 |
Hb_012347_010 |
0.1438689456 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |
| 6 |
Hb_003018_180 |
0.1487305017 |
- |
- |
PREDICTED: WEB family protein At3g51220-like [Jatropha curcas] |
| 7 |
Hb_027751_040 |
0.1565457243 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 8 |
Hb_000720_180 |
0.1575068896 |
- |
- |
- |
| 9 |
Hb_000957_040 |
0.162515356 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 10 |
Hb_124315_040 |
0.1661500434 |
- |
- |
hypothetical protein 19 [Hevea brasiliensis] |
| 11 |
Hb_010872_060 |
0.1679881813 |
transcription factor |
TF Family: MYB |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 12 |
Hb_001140_160 |
0.1746472435 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 13 |
Hb_000707_080 |
0.1784565155 |
- |
- |
hypothetical protein AALP_AA4G156500, partial [Arabis alpina] |
| 14 |
Hb_000762_020 |
0.1787393285 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 15 |
Hb_000002_390 |
0.1793036186 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_003756_010 |
0.1840692729 |
- |
- |
PREDICTED: uncharacterized protein LOC105642108 [Jatropha curcas] |
| 17 |
Hb_001780_060 |
0.1842876337 |
- |
- |
PREDICTED: uncharacterized protein LOC105631129 [Jatropha curcas] |
| 18 |
Hb_002027_300 |
0.1849195053 |
- |
- |
PREDICTED: rho GTPase-activating protein 2 [Jatropha curcas] |
| 19 |
Hb_000997_260 |
0.184997888 |
- |
- |
PREDICTED: rRNA-processing protein EFG1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000510_100 |
0.1951722682 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |