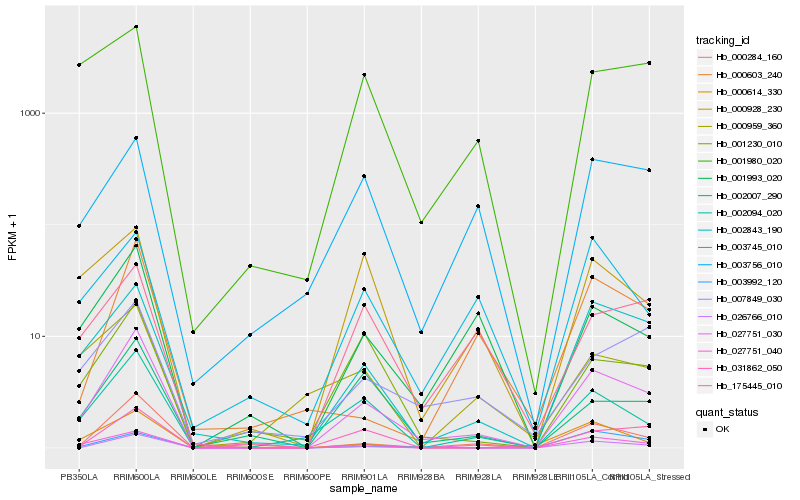
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027751_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_027751_040 |
0.1650707348 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 3 |
Hb_002843_190 |
0.1735290058 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 7 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002094_020 |
0.1793529519 |
- |
- |
PREDICTED: alpha-copaene synthase-like [Jatropha curcas] |
| 5 |
Hb_000284_160 |
0.1812820207 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 6 |
Hb_026766_010 |
0.1945522476 |
- |
- |
- |
| 7 |
Hb_001230_010 |
0.2214952176 |
- |
- |
- |
| 8 |
Hb_001993_020 |
0.2222613562 |
- |
- |
hypothetical protein JCGZ_02641 [Jatropha curcas] |
| 9 |
Hb_002007_290 |
0.2228280719 |
- |
- |
- |
| 10 |
Hb_007849_030 |
0.2346565866 |
- |
- |
l-allo-threonine aldolase, putative [Ricinus communis] |
| 11 |
Hb_000614_330 |
0.2391037443 |
- |
- |
hypothetical protein POPTR_0008s18070g [Populus trichocarpa] |
| 12 |
Hb_175445_010 |
0.2437165502 |
- |
- |
Pectin methylesterase 31 isoform 2, partial [Theobroma cacao] |
| 13 |
Hb_003756_010 |
0.2441010139 |
- |
- |
PREDICTED: uncharacterized protein LOC105642108 [Jatropha curcas] |
| 14 |
Hb_000603_240 |
0.2456577602 |
- |
- |
- |
| 15 |
Hb_003745_010 |
0.247682485 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |
| 16 |
Hb_000928_230 |
0.2488984984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001980_020 |
0.2505993466 |
- |
- |
- |
| 18 |
Hb_031862_050 |
0.251356459 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 19 |
Hb_003992_120 |
0.2540888344 |
- |
- |
hypothetical protein MTR_6g089560 [Medicago truncatula] |
| 20 |
Hb_000959_360 |
0.2598068265 |
- |
- |
transporter, putative [Ricinus communis] |