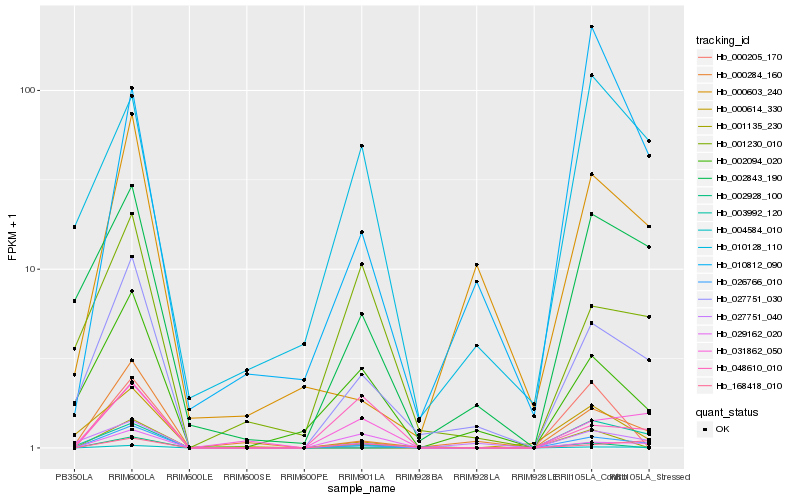
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026766_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_027751_030 |
0.1945522476 |
- |
- |
- |
| 3 |
Hb_000284_160 |
0.2002782405 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 4 |
Hb_003992_120 |
0.2440557866 |
- |
- |
hypothetical protein MTR_6g089560 [Medicago truncatula] |
| 5 |
Hb_004584_010 |
0.2570622468 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 6 |
Hb_000614_330 |
0.2582185364 |
- |
- |
hypothetical protein POPTR_0008s18070g [Populus trichocarpa] |
| 7 |
Hb_000603_240 |
0.2597243152 |
- |
- |
- |
| 8 |
Hb_002843_190 |
0.2603536147 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 7 isoform X1 [Jatropha curcas] |
| 9 |
Hb_031862_050 |
0.2677673438 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 10 |
Hb_027751_040 |
0.277738274 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 11 |
Hb_002094_020 |
0.2864139042 |
- |
- |
PREDICTED: alpha-copaene synthase-like [Jatropha curcas] |
| 12 |
Hb_002928_100 |
0.2883558023 |
- |
- |
- |
| 13 |
Hb_001135_230 |
0.2914578105 |
- |
- |
PREDICTED: protein REVEILLE 6-like isoform X3 [Gossypium raimondii] |
| 14 |
Hb_000205_170 |
0.3170332384 |
- |
- |
PREDICTED: uncharacterized protein LOC103926801 [Pyrus x bretschneideri] |
| 15 |
Hb_010128_110 |
0.320443925 |
- |
- |
PREDICTED: uncharacterized protein LOC105632071 [Jatropha curcas] |
| 16 |
Hb_010812_090 |
0.3211251396 |
- |
- |
PREDICTED: serine/threonine-protein kinase AtPK2/AtPK19-like [Jatropha curcas] |
| 17 |
Hb_168418_010 |
0.3224605211 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 18 |
Hb_029162_020 |
0.327320894 |
- |
- |
- |
| 19 |
Hb_048610_010 |
0.3307378514 |
- |
- |
- |
| 20 |
Hb_001230_010 |
0.3318274432 |
- |
- |
- |