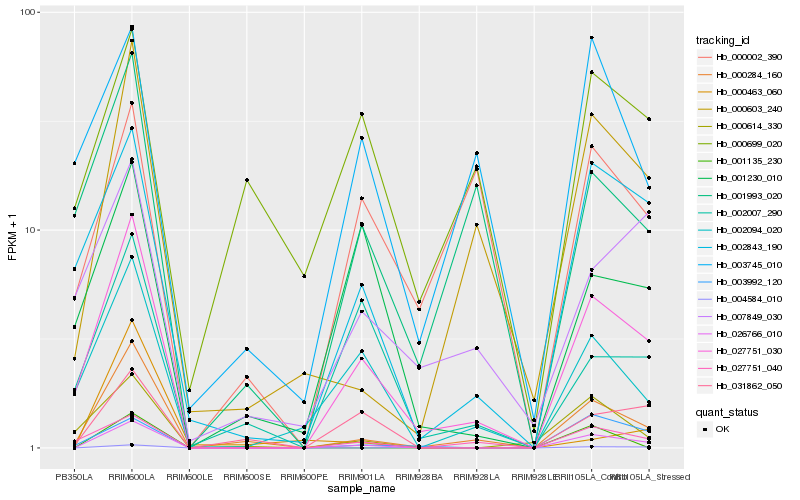
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000284_160 |
0.0 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 2 |
Hb_027751_030 |
0.1812820207 |
- |
- |
- |
| 3 |
Hb_000603_240 |
0.1881887859 |
- |
- |
- |
| 4 |
Hb_026766_010 |
0.2002782405 |
- |
- |
- |
| 5 |
Hb_027751_040 |
0.2455691889 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 6 |
Hb_000614_330 |
0.2578513535 |
- |
- |
hypothetical protein POPTR_0008s18070g [Populus trichocarpa] |
| 7 |
Hb_001993_020 |
0.2583102947 |
- |
- |
hypothetical protein JCGZ_02641 [Jatropha curcas] |
| 8 |
Hb_002094_020 |
0.2650565124 |
- |
- |
PREDICTED: alpha-copaene synthase-like [Jatropha curcas] |
| 9 |
Hb_002843_190 |
0.2792531846 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 7 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004584_010 |
0.2937247129 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 11 |
Hb_031862_050 |
0.2977342659 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 12 |
Hb_002007_290 |
0.3037110254 |
- |
- |
- |
| 13 |
Hb_003745_010 |
0.3080660299 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |
| 14 |
Hb_003992_120 |
0.3167867417 |
- |
- |
hypothetical protein MTR_6g089560 [Medicago truncatula] |
| 15 |
Hb_001135_230 |
0.3176606136 |
- |
- |
PREDICTED: protein REVEILLE 6-like isoform X3 [Gossypium raimondii] |
| 16 |
Hb_007849_030 |
0.3179243203 |
- |
- |
l-allo-threonine aldolase, putative [Ricinus communis] |
| 17 |
Hb_000463_060 |
0.3246623657 |
- |
- |
PREDICTED: uncharacterized protein LOC105638432 [Jatropha curcas] |
| 18 |
Hb_000699_020 |
0.3248830695 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |
| 19 |
Hb_001230_010 |
0.3293392092 |
- |
- |
- |
| 20 |
Hb_000002_390 |
0.3311827553 |
- |
- |
DNA binding protein, putative [Ricinus communis] |