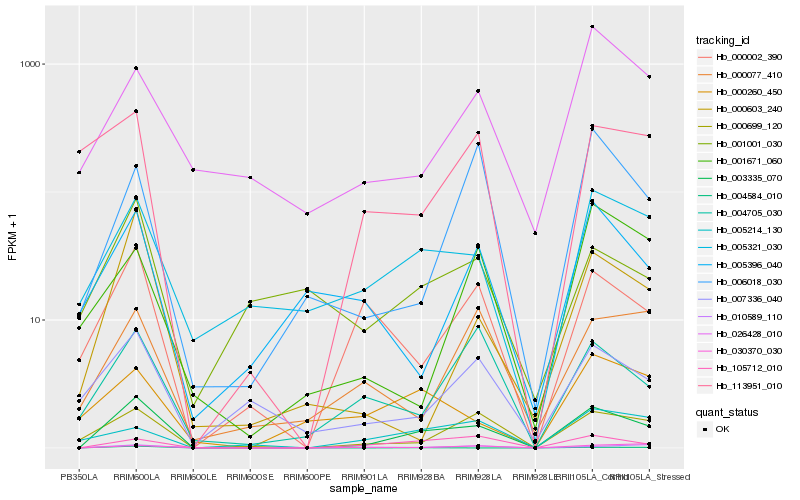
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003335_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_000699_120 |
0.2428665193 |
- |
- |
DNA-binding family protein [Populus trichocarpa] |
| 3 |
Hb_105712_010 |
0.2460860379 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 4 |
Hb_000603_240 |
0.2706962379 |
- |
- |
- |
| 5 |
Hb_000002_390 |
0.2772227763 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_010589_110 |
0.279234249 |
- |
- |
PREDICTED: pumilio homolog 12-like [Jatropha curcas] |
| 7 |
Hb_005321_030 |
0.2795833697 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4 [Jatropha curcas] |
| 8 |
Hb_004705_030 |
0.2866405691 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007336_040 |
0.2938799424 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_004584_010 |
0.294091362 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 11 |
Hb_006018_030 |
0.3009850696 |
- |
- |
PREDICTED: uncharacterized protein LOC105108875 isoform X1 [Populus euphratica] |
| 12 |
Hb_113951_010 |
0.3089742956 |
- |
- |
hypothetical protein JCGZ_06358 [Jatropha curcas] |
| 13 |
Hb_005214_130 |
0.3105733001 |
- |
- |
- |
| 14 |
Hb_000260_450 |
0.3129464002 |
- |
- |
GTP binding protein, putative [Ricinus communis] |
| 15 |
Hb_000077_410 |
0.3168897531 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_026428_010 |
0.3178728913 |
- |
- |
copper transport protein ATOX1 [Hevea brasiliensis] |
| 17 |
Hb_030370_030 |
0.317878885 |
- |
- |
putative wall-associated kinase family protein [Populus trichocarpa] |
| 18 |
Hb_005396_040 |
0.3224842395 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_001001_030 |
0.32518201 |
- |
- |
hypothetical protein POPTR_0014s03230g [Populus trichocarpa] |
| 20 |
Hb_001671_060 |
0.32622459 |
- |
- |
fad NAD binding oxidoreductases, putative [Ricinus communis] |