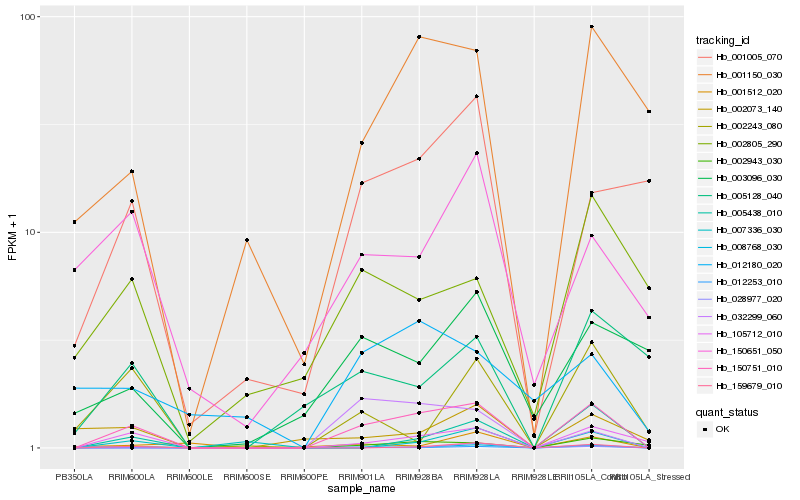
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_150751_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 2 |
Hb_105712_010 |
0.2354830819 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 3 |
Hb_012253_010 |
0.2669940047 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 4 |
Hb_002943_030 |
0.2679334986 |
- |
- |
- |
| 5 |
Hb_159679_010 |
0.3035112836 |
- |
- |
PREDICTED: uncharacterized protein LOC105786838 [Gossypium raimondii] |
| 6 |
Hb_008768_030 |
0.3046658042 |
- |
- |
- |
| 7 |
Hb_001150_030 |
0.3186058278 |
- |
- |
NADP-dependent oxidoreductase P1 isoform 3 [Theobroma cacao] |
| 8 |
Hb_005438_010 |
0.3223256729 |
- |
- |
PREDICTED: uncharacterized protein LOC104902779 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_001005_070 |
0.3317550609 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF113-like [Jatropha curcas] |
| 10 |
Hb_005128_040 |
0.3579196548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_150651_050 |
0.3609572965 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 12 |
Hb_003096_030 |
0.3633670318 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002243_080 |
0.3636165493 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Vitis vinifera] |
| 14 |
Hb_002073_140 |
0.364117205 |
- |
- |
- |
| 15 |
Hb_028977_020 |
0.3661252354 |
- |
- |
hypothetical protein JCGZ_21319 [Jatropha curcas] |
| 16 |
Hb_007336_030 |
0.3671874259 |
- |
- |
- |
| 17 |
Hb_032299_060 |
0.371824311 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC-like [Elaeis guineensis] |
| 18 |
Hb_002805_290 |
0.3754793899 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_001512_020 |
0.375623236 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |
| 20 |
Hb_012180_020 |
0.378108035 |
- |
- |
hypothetical protein POPTR_0001s15100g [Populus trichocarpa] |