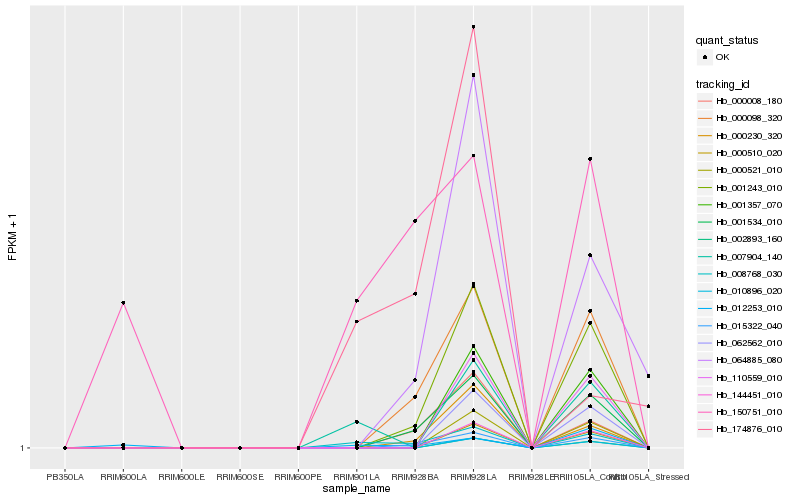
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008768_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_007904_140 |
0.2265411508 |
- |
- |
- |
| 3 |
Hb_015322_040 |
0.2513739055 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 4 |
Hb_001534_010 |
0.2518943119 |
- |
- |
Alpha-expansin 3 precursor [Theobroma cacao] |
| 5 |
Hb_000098_320 |
0.2563288692 |
- |
- |
- |
| 6 |
Hb_001243_010 |
0.2613713476 |
- |
- |
PREDICTED: dirigent protein 4-like [Populus euphratica] |
| 7 |
Hb_000008_180 |
0.2805249642 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000230_320 |
0.2851295799 |
- |
- |
PREDICTED: AP-5 complex subunit mu-like [Nelumbo nucifera] |
| 9 |
Hb_012253_010 |
0.2971114784 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 10 |
Hb_150751_010 |
0.3046658042 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 11 |
Hb_174876_010 |
0.3284671244 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 12 |
Hb_064885_080 |
0.3322933478 |
- |
- |
- |
| 13 |
Hb_000510_020 |
0.3453074599 |
- |
- |
PREDICTED: uncharacterized protein LOC103415345 [Malus domestica] |
| 14 |
Hb_144451_010 |
0.3453084194 |
- |
- |
hypothetical protein VITISV_001808 [Vitis vinifera] |
| 15 |
Hb_002893_160 |
0.3453174873 |
- |
- |
peptidase, putative [Ricinus communis] |
| 16 |
Hb_010896_020 |
0.3453179482 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_000521_010 |
0.345332025 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC [Jatropha curcas] |
| 18 |
Hb_062562_010 |
0.3454451185 |
- |
- |
PREDICTED: uncharacterized protein LOC104219776 [Nicotiana sylvestris] |
| 19 |
Hb_110559_010 |
0.3459236632 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 20 |
Hb_001357_070 |
0.3460579851 |
- |
- |
hypothetical protein CISIN_1g0004672mg, partial [Citrus sinensis] |