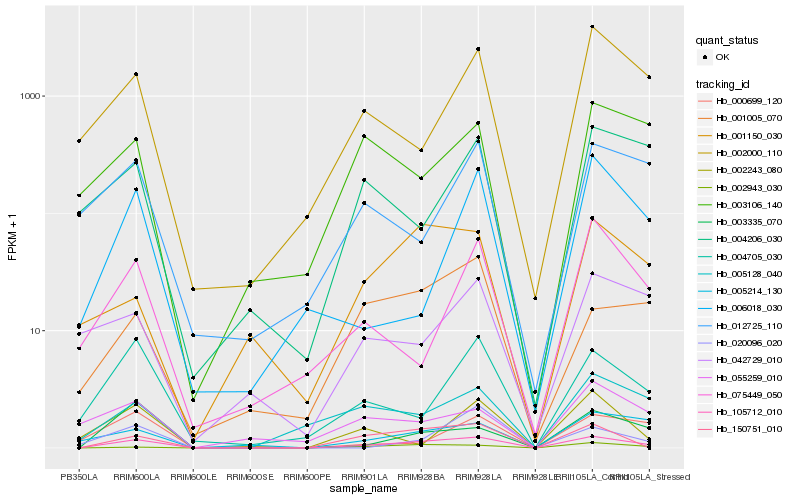
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_105712_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 2 |
Hb_002943_030 |
0.224292771 |
- |
- |
- |
| 3 |
Hb_150751_010 |
0.2354830819 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 4 |
Hb_003335_070 |
0.2460860379 |
- |
- |
- |
| 5 |
Hb_005128_040 |
0.2487424142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004705_030 |
0.2492294139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005214_130 |
0.2542736684 |
- |
- |
- |
| 8 |
Hb_002000_110 |
0.263305047 |
- |
- |
ASR-like protein 2 [Hevea brasiliensis] |
| 9 |
Hb_001005_070 |
0.2691213382 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF113-like [Jatropha curcas] |
| 10 |
Hb_001150_030 |
0.2691326676 |
- |
- |
NADP-dependent oxidoreductase P1 isoform 3 [Theobroma cacao] |
| 11 |
Hb_075449_050 |
0.2742623923 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 12 |
Hb_006018_030 |
0.2842174706 |
- |
- |
PREDICTED: uncharacterized protein LOC105108875 isoform X1 [Populus euphratica] |
| 13 |
Hb_012725_110 |
0.285955035 |
- |
- |
hypothetical protein JCGZ_09397 [Jatropha curcas] |
| 14 |
Hb_003106_140 |
0.2868128383 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000699_120 |
0.289814163 |
- |
- |
DNA-binding family protein [Populus trichocarpa] |
| 16 |
Hb_042729_010 |
0.2899798885 |
- |
- |
- |
| 17 |
Hb_055259_010 |
0.2900379147 |
- |
- |
- |
| 18 |
Hb_004206_030 |
0.2901763954 |
- |
- |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 19 |
Hb_002243_080 |
0.2903593716 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Vitis vinifera] |
| 20 |
Hb_020096_020 |
0.2916970739 |
- |
- |
- |