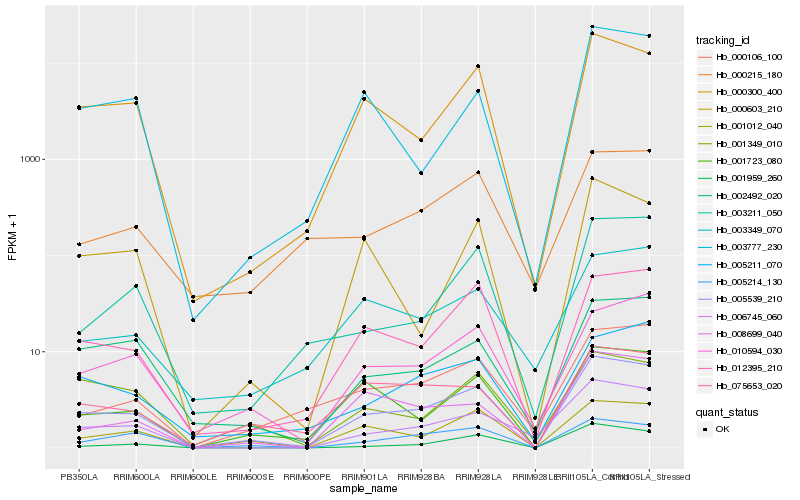
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005539_210 |
0.0 |
- |
- |
- |
| 2 |
Hb_000300_400 |
0.1136146124 |
- |
- |
hypothetical protein PHAVU_001G114100g [Phaseolus vulgaris] |
| 3 |
Hb_012395_210 |
0.132793512 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001012_040 |
0.1371047574 |
- |
- |
PREDICTED: MADS-box transcription factor 23-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_002492_020 |
0.1388486401 |
- |
- |
PREDICTED: uncharacterized protein LOC104789613 [Camelina sativa] |
| 6 |
Hb_010594_030 |
0.1401956269 |
- |
- |
- |
| 7 |
Hb_001723_080 |
0.1426762942 |
- |
- |
hypothetical protein L484_026139 [Morus notabilis] |
| 8 |
Hb_006745_060 |
0.1465374615 |
- |
- |
- |
| 9 |
Hb_075653_020 |
0.1482188189 |
- |
- |
- |
| 10 |
Hb_005211_070 |
0.1507579658 |
- |
- |
PREDICTED: uncharacterized protein LOC105642009 [Jatropha curcas] |
| 11 |
Hb_005214_130 |
0.1533369049 |
- |
- |
- |
| 12 |
Hb_001959_260 |
0.1582940376 |
- |
- |
- |
| 13 |
Hb_008699_040 |
0.1608960601 |
- |
- |
- |
| 14 |
Hb_000106_100 |
0.1649125103 |
- |
- |
- |
| 15 |
Hb_000603_210 |
0.1667980909 |
- |
- |
hypothetical protein POPTR_0006s23660g [Populus trichocarpa] |
| 16 |
Hb_003349_070 |
0.1698593471 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |
| 17 |
Hb_001349_010 |
0.1703126239 |
- |
- |
- |
| 18 |
Hb_003777_230 |
0.1703980338 |
- |
- |
PREDICTED: abscisic stress-ripening protein 1-like [Citrus sinensis] |
| 19 |
Hb_003211_050 |
0.1726005344 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 20 |
Hb_000215_180 |
0.1734356372 |
- |
- |
hypothetical protein 17 [Hevea brasiliensis] |