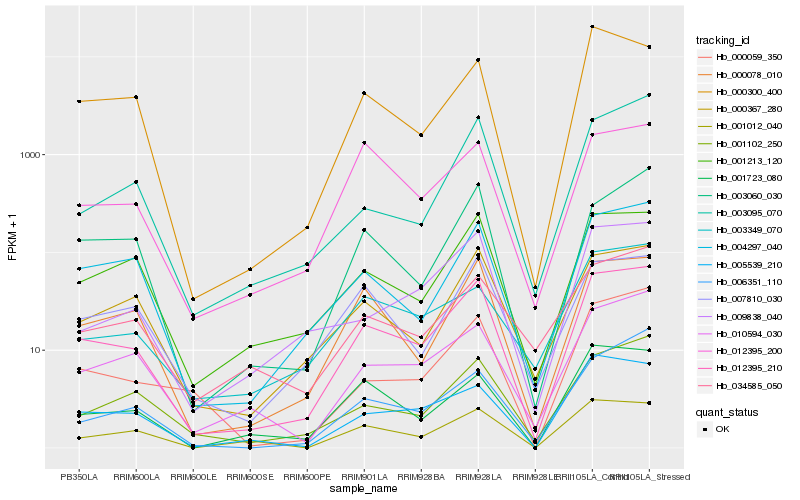
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012395_210 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004297_040 |
0.1187590131 |
- |
- |
acyl CoA reductase [Hevea brasiliensis] |
| 3 |
Hb_000078_010 |
0.1206600481 |
- |
- |
hypothetical protein POPTR_0006s06510g, partial [Populus trichocarpa] |
| 4 |
Hb_001213_120 |
0.1209458253 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Brassica rapa] |
| 5 |
Hb_000367_280 |
0.1284411812 |
- |
- |
PREDICTED: flavin-containing monooxygenase FMO GS-OX-like 9 [Jatropha curcas] |
| 6 |
Hb_001102_250 |
0.1303335647 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CCNB1IP1 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_005539_210 |
0.132793512 |
- |
- |
- |
| 8 |
Hb_010594_030 |
0.1338215556 |
- |
- |
- |
| 9 |
Hb_001012_040 |
0.1357939994 |
- |
- |
PREDICTED: MADS-box transcription factor 23-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_003060_030 |
0.1373660663 |
- |
- |
conserved hypothetical protein 16 [Hevea brasiliensis] |
| 11 |
Hb_000300_400 |
0.1375866538 |
- |
- |
hypothetical protein PHAVU_001G114100g [Phaseolus vulgaris] |
| 12 |
Hb_009838_040 |
0.1380655507 |
- |
- |
hypothetical protein JCGZ_12866 [Jatropha curcas] |
| 13 |
Hb_001723_080 |
0.1381618595 |
- |
- |
hypothetical protein L484_026139 [Morus notabilis] |
| 14 |
Hb_000059_350 |
0.1405589045 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_007810_030 |
0.1456300992 |
- |
- |
PREDICTED: uncharacterized protein LOC105647693 [Jatropha curcas] |
| 16 |
Hb_012395_200 |
0.147881564 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003095_070 |
0.1504843277 |
- |
- |
hypothetical protein POPTR_0009s11230g [Populus trichocarpa] |
| 18 |
Hb_034585_050 |
0.1518298019 |
- |
- |
- |
| 19 |
Hb_006351_110 |
0.1544062881 |
- |
- |
- |
| 20 |
Hb_003349_070 |
0.1576135593 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |