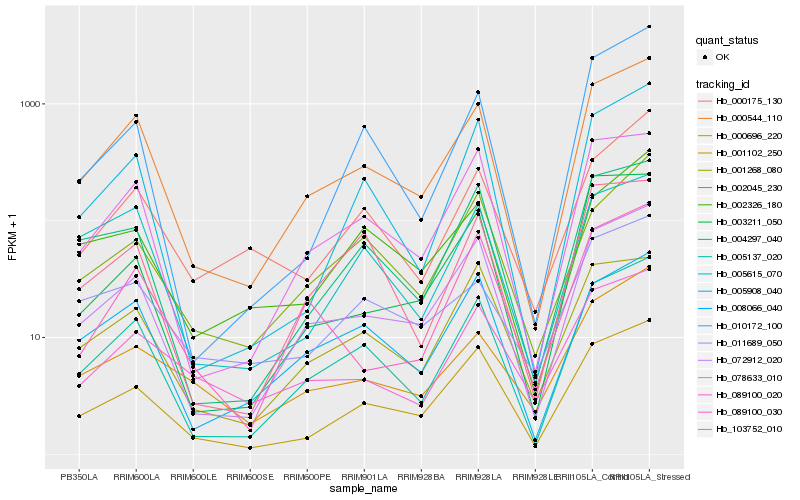
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000544_110 |
0.0 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 2 |
Hb_005137_020 |
0.0530811973 |
- |
- |
PREDICTED: uncharacterized protein LOC101759641 [Setaria italica] |
| 3 |
Hb_072912_020 |
0.0693488272 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 4 |
Hb_001102_250 |
0.097931815 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CCNB1IP1 homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_078633_010 |
0.0996783776 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 6 |
Hb_008066_040 |
0.1172258108 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL62 [Jatropha curcas] |
| 7 |
Hb_005908_040 |
0.118337121 |
- |
- |
hypothetical protein POPTR_0017s06850g [Populus trichocarpa] |
| 8 |
Hb_004297_040 |
0.1183809411 |
- |
- |
acyl CoA reductase [Hevea brasiliensis] |
| 9 |
Hb_001268_080 |
0.1338017257 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_005615_070 |
0.1381357727 |
- |
- |
Rab7 [Hevea brasiliensis] |
| 11 |
Hb_089100_020 |
0.1396527456 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 12 |
Hb_000175_130 |
0.1406209394 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase At3g15890 [Jatropha curcas] |
| 13 |
Hb_103752_010 |
0.1407951785 |
- |
- |
PREDICTED: uncharacterized protein At5g39865-like [Jatropha curcas] |
| 14 |
Hb_002326_180 |
0.1408323124 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 15 |
Hb_002045_230 |
0.1410337253 |
- |
- |
Ammonium transporter 2 [Theobroma cacao] |
| 16 |
Hb_000696_220 |
0.14214138 |
- |
- |
PREDICTED: uncharacterized protein LOC105647113 [Jatropha curcas] |
| 17 |
Hb_003211_050 |
0.1430541679 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 18 |
Hb_089100_030 |
0.1437656036 |
- |
- |
- |
| 19 |
Hb_011689_050 |
0.1445488933 |
- |
- |
hypothetical protein POPTR_0008s20290g [Populus trichocarpa] |
| 20 |
Hb_010172_100 |
0.14456081 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Populus euphratica] |