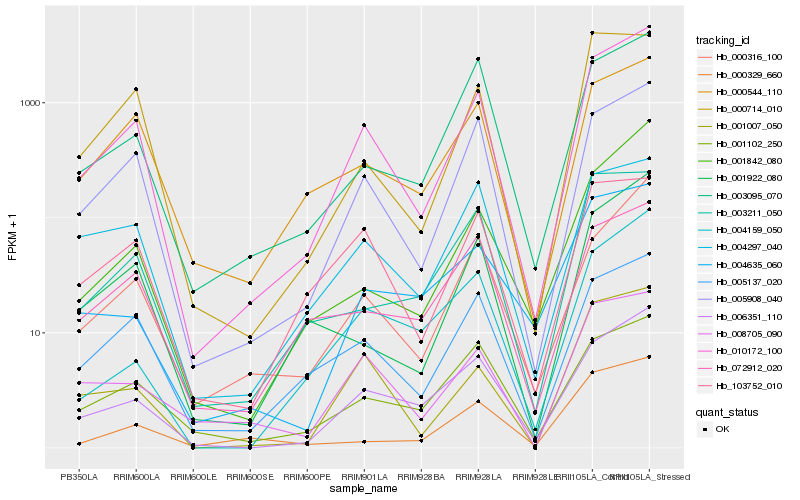
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010172_100 |
0.0 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Populus euphratica] |
| 2 |
Hb_005908_040 |
0.0991296418 |
- |
- |
hypothetical protein POPTR_0017s06850g [Populus trichocarpa] |
| 3 |
Hb_006351_110 |
0.1080985756 |
- |
- |
- |
| 4 |
Hb_005137_020 |
0.1252291139 |
- |
- |
PREDICTED: uncharacterized protein LOC101759641 [Setaria italica] |
| 5 |
Hb_000316_100 |
0.126608985 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_001007_050 |
0.1281565749 |
- |
- |
hypothetical protein MIMGU_mgv1a0196712mg, partial [Erythranthe guttata] |
| 7 |
Hb_001922_080 |
0.1415685512 |
- |
- |
Tellurite resistance protein tehA, putative [Ricinus communis] |
| 8 |
Hb_008705_090 |
0.1427874442 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2-like [Jatropha curcas] |
| 9 |
Hb_000544_110 |
0.14456081 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 10 |
Hb_001842_080 |
0.1484436482 |
- |
- |
- |
| 11 |
Hb_001102_250 |
0.1503598067 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CCNB1IP1 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_003095_070 |
0.1514756542 |
- |
- |
hypothetical protein POPTR_0009s11230g [Populus trichocarpa] |
| 13 |
Hb_000714_010 |
0.1548050792 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 14 |
Hb_004159_050 |
0.1578679023 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_103752_010 |
0.1613415571 |
- |
- |
PREDICTED: uncharacterized protein At5g39865-like [Jatropha curcas] |
| 16 |
Hb_003211_050 |
0.1620280972 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 17 |
Hb_072912_020 |
0.1624573264 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 18 |
Hb_004297_040 |
0.1633275105 |
- |
- |
acyl CoA reductase [Hevea brasiliensis] |
| 19 |
Hb_000329_660 |
0.1664435042 |
- |
- |
PREDICTED: carotenoid cleavage dioxygenase 7, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004635_060 |
0.1675091763 |
- |
- |
hypothetical protein POPTR_0006s22850g [Populus trichocarpa] |