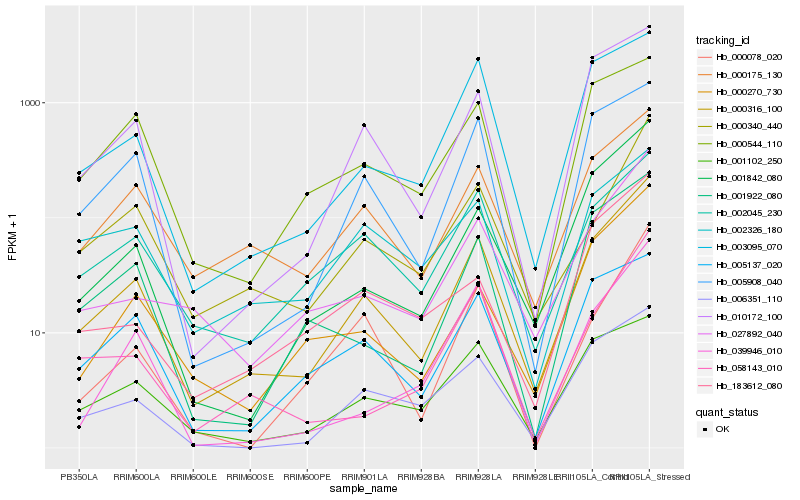
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000316_100 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_000175_130 |
0.1243018661 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase At3g15890 [Jatropha curcas] |
| 3 |
Hb_010172_100 |
0.126608985 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Populus euphratica] |
| 4 |
Hb_001842_080 |
0.1326632138 |
- |
- |
- |
| 5 |
Hb_006351_110 |
0.1350253087 |
- |
- |
- |
| 6 |
Hb_005908_040 |
0.1359348035 |
- |
- |
hypothetical protein POPTR_0017s06850g [Populus trichocarpa] |
| 7 |
Hb_027892_040 |
0.1362315771 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 8 |
Hb_002045_230 |
0.1422740987 |
- |
- |
Ammonium transporter 2 [Theobroma cacao] |
| 9 |
Hb_001922_080 |
0.1449570658 |
- |
- |
Tellurite resistance protein tehA, putative [Ricinus communis] |
| 10 |
Hb_000340_440 |
0.1453595043 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 11 |
Hb_003095_070 |
0.1461267809 |
- |
- |
hypothetical protein POPTR_0009s11230g [Populus trichocarpa] |
| 12 |
Hb_000270_730 |
0.1464848711 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa-like [Jatropha curcas] |
| 13 |
Hb_058143_010 |
0.1557463181 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 14 |
Hb_005137_020 |
0.1560794537 |
- |
- |
PREDICTED: uncharacterized protein LOC101759641 [Setaria italica] |
| 15 |
Hb_001102_250 |
0.157781339 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CCNB1IP1 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_000078_020 |
0.1578148389 |
- |
- |
- |
| 17 |
Hb_039946_010 |
0.1647674001 |
- |
- |
hypothetical protein POPTR_0018s09200g [Populus trichocarpa] |
| 18 |
Hb_000544_110 |
0.1679556354 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 19 |
Hb_002326_180 |
0.1682174285 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 20 |
Hb_183612_080 |
0.169847755 |
- |
- |
hypothetical protein POPTR_0001s00500g [Populus trichocarpa] |