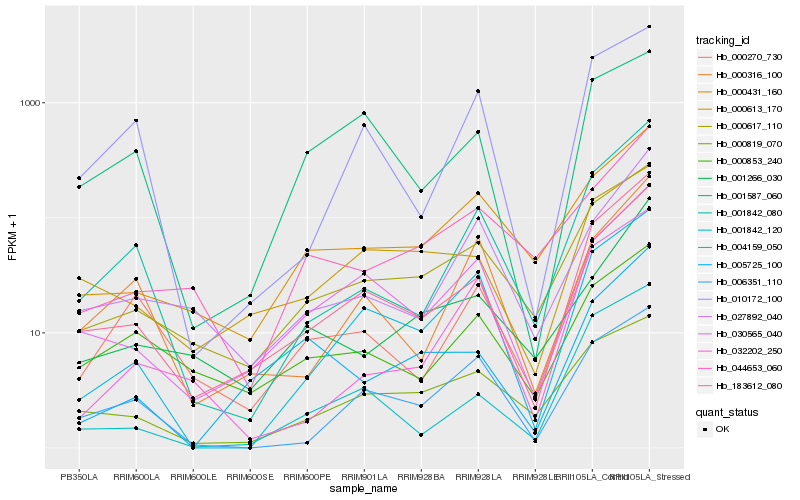
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183612_080 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s00500g [Populus trichocarpa] |
| 2 |
Hb_000617_110 |
0.119681007 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 3 |
Hb_000270_730 |
0.1307616732 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa-like [Jatropha curcas] |
| 4 |
Hb_001842_120 |
0.1408904811 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 5 |
Hb_001842_080 |
0.1418701177 |
- |
- |
- |
| 6 |
Hb_004159_050 |
0.1486941135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000613_170 |
0.1504113775 |
- |
- |
PREDICTED: RPM1-interacting protein 4 [Jatropha curcas] |
| 8 |
Hb_000431_160 |
0.1586087174 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 9 |
Hb_027892_040 |
0.1594706822 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 10 |
Hb_000316_100 |
0.169847755 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_000853_240 |
0.1710066271 |
- |
- |
- |
| 12 |
Hb_006351_110 |
0.1716326512 |
- |
- |
- |
| 13 |
Hb_001266_030 |
0.1718691637 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_000819_070 |
0.1720579038 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 1 [Jatropha curcas] |
| 15 |
Hb_005725_100 |
0.1736503083 |
- |
- |
hypothetical protein 30 [Hevea brasiliensis] |
| 16 |
Hb_010172_100 |
0.1755727524 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Populus euphratica] |
| 17 |
Hb_001587_060 |
0.1779349254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_044653_060 |
0.1798145041 |
- |
- |
PREDICTED: uncharacterized protein LOC105628750 [Jatropha curcas] |
| 19 |
Hb_032202_250 |
0.1814989901 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 20 |
Hb_030565_040 |
0.1815978948 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |