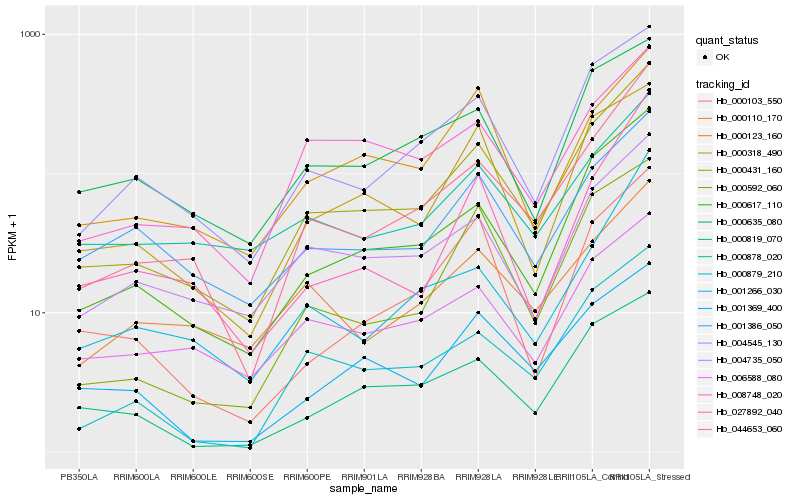
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000431_160 |
0.0 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 2 |
Hb_000617_110 |
0.0695259423 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 3 |
Hb_044653_060 |
0.0753166663 |
- |
- |
PREDICTED: uncharacterized protein LOC105628750 [Jatropha curcas] |
| 4 |
Hb_004545_130 |
0.0983965513 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 5 |
Hb_000879_210 |
0.1010711353 |
- |
- |
hypothetical protein POPTR_0014s01620g [Populus trichocarpa] |
| 6 |
Hb_004735_050 |
0.1165902658 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000819_070 |
0.1206219176 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 1 [Jatropha curcas] |
| 8 |
Hb_000318_490 |
0.1258045765 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM6 homolog [Jatropha curcas] |
| 9 |
Hb_000592_060 |
0.1275324865 |
- |
- |
PREDICTED: ras-related protein RABA6a [Populus euphratica] |
| 10 |
Hb_001369_400 |
0.1294018256 |
- |
- |
PREDICTED: uncharacterized protein LOC105648609 [Jatropha curcas] |
| 11 |
Hb_006588_080 |
0.1311440484 |
- |
- |
hypothetical protein JCGZ_25746 [Jatropha curcas] |
| 12 |
Hb_008748_020 |
0.1314148946 |
- |
- |
hypothetical protein B456_005G210700 [Gossypium raimondii] |
| 13 |
Hb_000123_160 |
0.1316860511 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 14 |
Hb_000635_080 |
0.1326280721 |
- |
- |
60S ribosomal protein L17A [Hevea brasiliensis] |
| 15 |
Hb_001386_050 |
0.1346369183 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 16 |
Hb_001266_030 |
0.1347055045 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_027892_040 |
0.1357741831 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 18 |
Hb_000878_020 |
0.137191596 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 19 |
Hb_000103_550 |
0.1376641017 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000110_170 |
0.1387895879 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |