| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
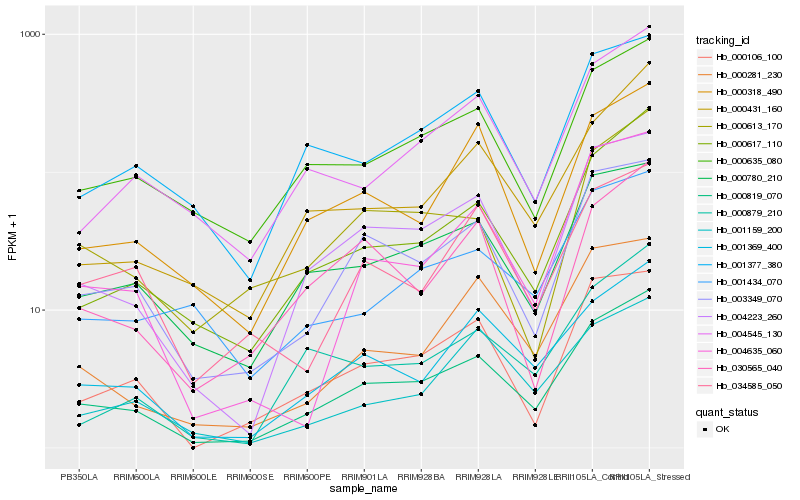
Hb_000819_070 |
0.0 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 1 [Jatropha curcas] |
| 2 |
Hb_004223_260 |
0.0863519553 |
- |
- |
hypothetical protein F383_09277 [Gossypium arboreum] |
| 3 |
Hb_000617_110 |
0.0988839968 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 4 |
Hb_001369_400 |
0.1004549612 |
- |
- |
PREDICTED: uncharacterized protein LOC105648609 [Jatropha curcas] |
| 5 |
Hb_003349_070 |
0.102603015 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |
| 6 |
Hb_000780_210 |
0.1121065707 |
- |
- |
PREDICTED: transmembrane protein 256 homolog [Jatropha curcas] |
| 7 |
Hb_000635_080 |
0.1121412883 |
- |
- |
60S ribosomal protein L17A [Hevea brasiliensis] |
| 8 |
Hb_000431_160 |
0.1206219176 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 9 |
Hb_000106_100 |
0.1226767422 |
- |
- |
- |
| 10 |
Hb_004545_130 |
0.1238531239 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 11 |
Hb_000318_490 |
0.1256976706 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM6 homolog [Jatropha curcas] |
| 12 |
Hb_001377_380 |
0.1272804023 |
- |
- |
PREDICTED: uncharacterized protein LOC105642831 [Jatropha curcas] |
| 13 |
Hb_004635_060 |
0.1313543309 |
- |
- |
hypothetical protein POPTR_0006s22850g [Populus trichocarpa] |
| 14 |
Hb_000281_230 |
0.1316409264 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 15 |
Hb_000613_170 |
0.1361381975 |
- |
- |
PREDICTED: RPM1-interacting protein 4 [Jatropha curcas] |
| 16 |
Hb_000879_210 |
0.1371640087 |
- |
- |
hypothetical protein POPTR_0014s01620g [Populus trichocarpa] |
| 17 |
Hb_001434_070 |
0.1406824081 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM8 [Jatropha curcas] |
| 18 |
Hb_001159_200 |
0.1413210961 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_030565_040 |
0.1420396192 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 20 |
Hb_034585_050 |
0.1427216319 |
- |
- |
- |