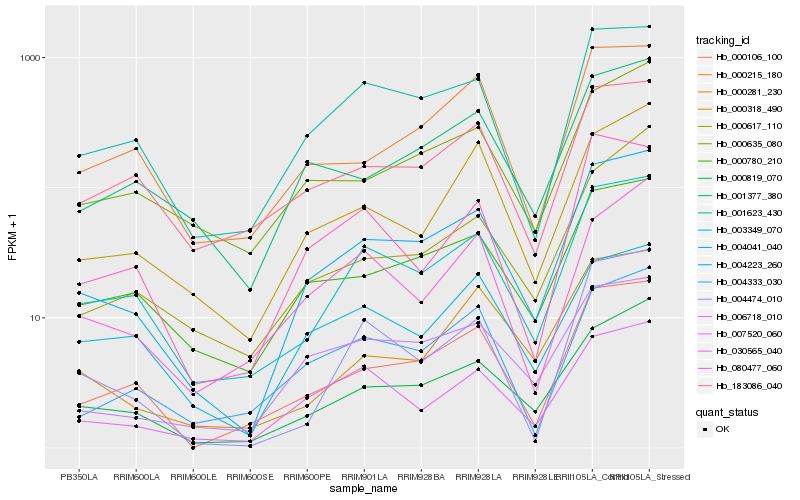
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004223_260 |
0.0 |
- |
- |
hypothetical protein F383_09277 [Gossypium arboreum] |
| 2 |
Hb_000819_070 |
0.0863519553 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 1 [Jatropha curcas] |
| 3 |
Hb_003349_070 |
0.0989922374 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |
| 4 |
Hb_000106_100 |
0.1052160056 |
- |
- |
- |
| 5 |
Hb_000780_210 |
0.1094336712 |
- |
- |
PREDICTED: transmembrane protein 256 homolog [Jatropha curcas] |
| 6 |
Hb_006718_010 |
0.1133044049 |
- |
- |
PREDICTED: histone H2AX-like [Elaeis guineensis] |
| 7 |
Hb_001623_430 |
0.1156731898 |
- |
- |
PREDICTED: small acidic protein 1 [Jatropha curcas] |
| 8 |
Hb_007520_060 |
0.1211055799 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |
| 9 |
Hb_001377_380 |
0.1242880033 |
- |
- |
PREDICTED: uncharacterized protein LOC105642831 [Jatropha curcas] |
| 10 |
Hb_000318_490 |
0.1276416423 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM6 homolog [Jatropha curcas] |
| 11 |
Hb_000635_080 |
0.1329525268 |
- |
- |
60S ribosomal protein L17A [Hevea brasiliensis] |
| 12 |
Hb_004333_030 |
0.1331361951 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 26A [Jatropha curcas] |
| 13 |
Hb_004474_010 |
0.1375039173 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_000281_230 |
0.1377112712 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 15 |
Hb_000215_180 |
0.1407017421 |
- |
- |
hypothetical protein 17 [Hevea brasiliensis] |
| 16 |
Hb_080477_060 |
0.1412008009 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000617_110 |
0.1415439283 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 18 |
Hb_030565_040 |
0.1445019541 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 19 |
Hb_004041_040 |
0.1456671165 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM14-1 [Camelina sativa] |
| 20 |
Hb_183086_040 |
0.1497584801 |
- |
- |
cytochrome b5 isoform Cb5-A [Vernicia fordii] |