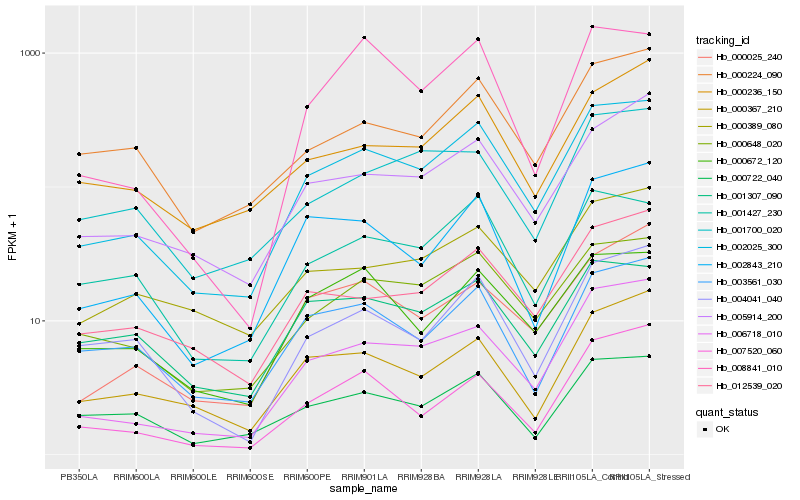
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002025_300 |
0.0 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 [Jatropha curcas] |
| 2 |
Hb_000648_020 |
0.0807735276 |
- |
- |
PREDICTED: chaperone protein dnaJ 10-like [Jatropha curcas] |
| 3 |
Hb_006718_010 |
0.0897870333 |
- |
- |
PREDICTED: histone H2AX-like [Elaeis guineensis] |
| 4 |
Hb_005914_200 |
0.1027738336 |
- |
- |
PREDICTED: 60S ribosomal protein L23-like isoform X1 [Tarenaya hassleriana] |
| 5 |
Hb_002843_210 |
0.1055341675 |
- |
- |
PREDICTED: uncharacterized protein LOC105647178 [Jatropha curcas] |
| 6 |
Hb_012539_020 |
0.1069342103 |
- |
- |
protein binding/ubiquitin-protein ligase 4 [Hevea brasiliensis] |
| 7 |
Hb_000025_240 |
0.1116200442 |
- |
- |
PREDICTED: uncharacterized protein LOC105628964 [Jatropha curcas] |
| 8 |
Hb_001427_230 |
0.1117260357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000672_120 |
0.1121932669 |
- |
- |
V-type proton ATPase subunit G 1 [Jatropha curcas] |
| 10 |
Hb_000224_090 |
0.1145418696 |
- |
- |
Eukaryotic translation initiation factor 1A [Gossypium arboreum] |
| 11 |
Hb_003561_030 |
0.1175991167 |
- |
- |
PREDICTED: uncharacterized protein LOC105645274 [Jatropha curcas] |
| 12 |
Hb_004041_040 |
0.1181282349 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM14-1 [Camelina sativa] |
| 13 |
Hb_000389_080 |
0.118343408 |
- |
- |
PREDICTED: uncharacterized protein At4g26485-like [Jatropha curcas] |
| 14 |
Hb_000236_150 |
0.1211509422 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 15 |
Hb_000722_040 |
0.1216551068 |
- |
- |
- |
| 16 |
Hb_000367_210 |
0.1223906754 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 3 [Jatropha curcas] |
| 17 |
Hb_001307_090 |
0.1245626944 |
- |
- |
hypothetical protein CISIN_1g0263261mg [Citrus sinensis] |
| 18 |
Hb_007520_060 |
0.1282422533 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |
| 19 |
Hb_001700_020 |
0.1288113147 |
- |
- |
PREDICTED: 60S ribosomal protein L37-1 [Jatropha curcas] |
| 20 |
Hb_008841_010 |
0.1294888688 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |